Open is not enough. Let’s take the next step: an integrated, community-driven computing platform for neuroscience
- 1 Center for Cognitive Neuroscience, Dartmouth College, Hanover, NH, USA
- 2 Department of Psychological and Brain Sciences, Dartmouth College, Hanover, NH, USA
- 3 Debian Project, http://www.debian.org
- 4 Department of Experimental Psychology, Otto-von-Guericke University, Magdeburg, Germany
- 5 Center for Behavioral Brain Sciences, Magdeburg, Germany
The last 5 years have seen dramatic improvements in the collaborative research infrastructure. A need for open research tools has been identified (Ince et al., 2012), and one solution has been clearing houses, such as the INCF Software Center1, and the NITRC2 portal, which facilitate efforts of peer-to-peer software and data sharing that were previously limited to only well-funded formal consortia (see, Poline et al., 2012, for a recent summary of the status quo). However, collecting these resources into a centralized clearing-house addresses only one necessary aspect on the way to a sustainable software ecosystem for neuroscience – availability. Unfortunately it does not ensure ease of deployment, nor does it offer a sustainable model for long-term maintenance.
At the same time, the development model of many neuroscience research software projects is broken. Inefficient and opaque procedures combined with a scarce developer workforce result in tools of insufficient quality and robustness that we rely on to conduct our research. Moreover, as the scientists, students, and research groups responsible for these tools move on to new tasks, their software is often left in a state of limbo, with no continued support for bug fixes or sufficiently coordinated maintenance. Over time, changes in underlying computing environments break the tools completely, and they commonly become abandoned – with costly consequences for the scientists depending upon them.
To address this problem, we need to bring our tools further into the open, and consolidate development efforts on an open and community-driven platform – one that is capable of providing easy access, installation, and maintenance for any research software. Such effort will not only help to improve aspects of software engineering, but also meet many unfulfilled requirements toward the goal of practical open science.
To the best of our knowledge, NeuroDebian3 is the most comprehensive attempt at improving the neuroscience software ecosystem. It applies proven principles and procedures from Free and Open-Source Software (FOSS) development to the maintenance and deployment of neuroscience software. NeuroDebian’s strategy is to work with scientists and developers, helping them to directly integrate their software with the Debian4 operating system (OS). Using the Debian OS as a foundation for these efforts offers five unique advantages for end-users and developers alike.
Installing and upgrading software takes only minutes – as a result of using the included software package management tools – regardless of application complexity. Knowledge of custom installation procedures is simply not needed. A recent survey confirms that researchers who use software package management systems spend less time on system administration and more time on scientifically relevant tasks (Hanke and Halchenko, 2011).
Strict open standards – for included software, as defined in Debian’s policy (Jackson et al., 2012), help yield the maximum interoperability of all components without sacrificing robustness (Fortuna et al., 2011) or creating unnecessary burdens. This has enabled developers to make routine releases of the Debian OS for the past 20 years, which is accomplished by a group of loosely connected individuals working together over the internet – not unlike collaborating scientists. Applying these established open standards to neuroscience software improves the longevity and robustness of open science.
Debian offers more than 29,000 readily usable pieces of software – making it the largest archive of its kind – especially for scientific purposes5. This wide selection is a priceless resource not only for using, but also for building atop these libraries and applications from a variety of fields of endeavor. The joint maintenance of shared components reduces the overall maintenance cost, (Ghosh, 2006) and makes the integration of special-interest neuroscience software both more time- and effort-effective.
Debian is open to anyone – Anyone is free to contribute anything to Debian6 within the limits set by its Social Contract (Perens et al., 1997). Debian is a well established self-governing project (O’Mahony and Ferraro, 2007) and is not controlled by any external entity that might impose a marketing strategy or decide what software ought to be eligible for inclusion into the system. Although the project is primarily focused on the development of an operating system built entirely from FOSS, the Social Contract explicitly acknowledges the necessity to also support restrictively licensed and proprietary closed-source solutions. Consequently, Debian provides all required tools and infrastructure to build and distribute non-free software as well. Debian’s “do-ocracy” principle puts the person(s) contributing completely in charge of the details of their contribution. This aspect allows for integration of any software directly into the Debian system, while also being able to rely on its existing infrastructure (bug tracking system, software repositories with more than a hundred mirrors worldwide, etc.). As a result, this largely abolishes the need for a research-specific software distribution infrastructure.
Standardization of binary and source distributions promotes reproducibility – both for troubleshooting and results. Additionally, standardized source packages encourage contributions by other developers – even ones who may be unfamiliar with the specific purpose of a piece of software. This helps to mitigate the shortage of man-power for research software maintenance.
We believe that these advantages make the Debian software ecosystem an ideal environment for consolidating neuroscience research software. The Debian OS has been ported to numerous hardware architectures and runs natively on any hardware relevant for neuroscience research. Additionally, through means of virtualization an entire system can be deployed in cloud-computing scenarios7 or run alongside commercial OS’s (e.g., OS X and MS Windows), thus making neuroscience software universally available.
For the past 6 years, NeuroDebian has consistently proven the validity of these points, especially regarding the efficiency of the Debian platform. NeuroDebian is largely the result of two researchers working in their spare time, with the help of a handful of additional volunteers8. Despite never having received direct institutional funding and having only limited man-power, today NeuroDebian software packages are used by thousands of researchers in hundreds of labs world-wide9 – with a current growth of about 20 new subscriptions each day (Figure 1). What had originally started as a small project to deploy neuroimaging-related software for personal convenience, now provides software for many related disciplines – electrophysiology, neural modeling, psychophysics, and distributed computing – thus enhancing its utility for multi-modal or multi-disciplinary projects. Over 50 packages have successfully migrated from NeuroDebian into Debian, which are now officially part of the Debian OS and its more than 10010 derived GNU/Linux distributions, such as Ubuntu11 (developed by Canonical Ltd.). By virtue of collaboration with other related efforts inside the Debian project (Debian Med12 and Debian Science13), Debian offers an impressive set of several hundred special-purpose tools of relevance for neuroscience research computing environments. As a result, the 2011 release of Debian 6.0 became the first OS to offer comprehensive support for neuroimaging research14. Moreover, the upcoming release of Debian 7.0 will nearly double the number of neuroscience-related packages already included in Debian.
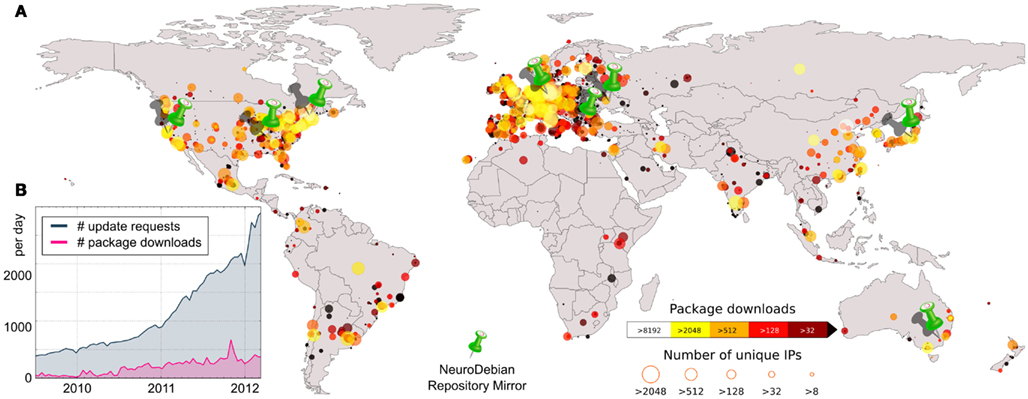
Figure 1. NeuroDebian repository access statistics from March 2009 to April 2012. (A) Geographic distribution of NeuroDebian repository accesses, aggregated by city or region as determined by GeoIP. Where no region-specific GeoIP information is available, statistics are placed into the center of the respective country. The color of each circle depicts the total number of package downloads and its size corresponds to the number of IP addresses which accessed the repository from that region. Push-pin icons indicate the location of NeuroDebian repository mirrors. (B) Daily statistic. The blue curve (# of update requests) shows the number of IP-address/OS-release combinations accessing the repository on any given day. This number should well approximate the lower bound of current “subscribers” of the repository. The red curve corresponds to the number of daily binary package downloads.
To complement these “official” packages, NeuroDebian provides a large number of additional software packages in a supplementary repository. This includes software which is still in the integration process, as well as “backports” of recent research software releases for older versions of Debian and Ubuntu – addressing researchers’ desire to combine the latest software technology with a stable computing platform. The NeuroDebian repository network (eight community-provided mirrors in six countries; see Figure 1) also distributes various data packages that are frequently used in research or teaching activities, such as stereotaxic atlases and example datasets15.
Apart from individual packages, NeuroDebian offers a complete Virtual Machine (VM) that can be used on any major operating system. VMs are a popular approach for streamlining the deployment of complex research software on less science-savvy platforms (Hanke and Halchenko, 2011), and to facilitate collaborative work through the availability of a common computing environment (see the strategy of the 2nd winner of “The Executable Paper Grand Challenge” by Elsevier, Gorp and Mazanek, 2011). Instead of being tuned for a single tool, such as VMs offered by individual research groups for their own particular software, the NeuroDebian VM is designed to offer a multitude of integrated research software. The NeuroDebian VM has been downloaded more than 2900 times, and is frequently used for educational workshops. Students are provided with a fully functional computing environment, which they can explore and take home alongside other course materials after the workshop or seminar has ended16. With such versatility, NeuroDebian offers a reliable foundation for day-to-day research activities. One such indicator for the robustness of the (Neuro)Debian system can be seen from the growing number of derivative works that are being built upon it17. To our knowledge, NeuroDebian serves as the basis for several virtual appliances (e.g., XNAT; Marcus et al., 2007), the Lin4Neuro distribution (Nemoto et al., 2011), as well as the NITRC Computational Environment18.
NeuroDebian has shown that by adopting established tools and workflows it is possible to create a powerful integrated environment with minimal investment – offering a practical solution that is compliant with all proposed requirements for transparency of research software (Morin et al., 2012). Our vision for the future of neuroscience research software is one where the whole scientific community fully embraces the principles of the FOSS movement. This will accelerate the dissemination of new software technologies, foster efforts to improve software quality, and facilitate reproducible research – eventually leading to a universally available, truly comprehensive, and sustainable integrated computing platform. To achieve this goal, it is necessary to distribute the work in a way that does not represent an additional and unnecessary burden for the individual scientist: (1) Developers of research software need to encourage and facilitate external contributions. This requires public access to all of the standard tools of a typical FOSS development workflow, such as a version control system, bug tracker, and mailing list. Software needs to be released under a standard FOSS license in order to avoid wasteful reimplementation efforts caused by license incompatibilities. Efforts to integrate individual tools into larger analysis suites or software distributions must be promoted19, as increased exposure by frequent re-use (instead of duplication) helps to improve software quality and minimize the “cost.” (2) Users of research software need to offer as much feedback as possible. Software authors need to know how, and how often, their tools are being used in order to be able to obtain sufficient funding for future development and maintenance. Knowledge of typical usage patterns is also critical to decide which functionality is most important and where development resources should be focused. There are tools to report anonymous usage statistics automatically (e.g., Debian popularity contest20), abolishing the need for time-consuming manual actions. When software defects are discovered, they must be reported immediately. Reports need to be directed to a public forum to allow other researchers to check for the status of a particular defect and coordinate the efforts to fix it. Engagement with end-users will help guarantee the robust and correct operation of their computing research environments and thus improve the quality and productivity of research.
While these actions only require very little time from an individual researcher, their combined effect when adopted by a community can be massive. The breath-taking development pace of the NiPy21 and NeuralEnsemble22 communities are flagship examples of the power and potential of what can be achieved when the proposed principles are put into effect.
As a global community of scientists, it is now our chance to take the next step toward an accessible open science. Together we can create an integrated computing platform that we all freely share, to exchange data and ideas, implemented as software, that we all maintain collaboratively. It is unlikely that any single project with a classical funding scheme will ever accomplish what is needed. The tools are there – we just need to do it.
Acknowledgments
We are grateful to all Debian contributors for their dedication to free software. Thanks to the developers of open research software which make NeuroDebian possible and “open science” feasible. We are indebted to our colleagues for their critical comments, and to Alex Waite for his work on improving the readability of this manuscript. Lastly, our sincere gratitude is extended to Jim Haxby for his generous and enduring support of NeuroDebian.
Footnotes
- ^http://software.incf.org
- ^http://www.nitrc.org
- ^http://neuro.debian.net
- ^http://www.debian.org
- ^http://blends.alioth.debian.org/science/tasks
- ^http://mentors.debian.net provides a convenient gateway for contributions from the community
- ^http://wiki.debian.org/Cloud/AmazonEC2Image
- ^http://neuro.debian.net/pkgs.html#by-maintainer for an up-to-date list of contributors
- ^http://neuro.debian.net/testimonials.html
- ^http://distrowatch.com/search.php?basedon=Debian
- ^www.ubuntu.com
- ^http://wiki.debian.org/DebianMed
- ^http://wiki.debian.org/DebianScience
- ^http://www.debian.org/releases/stable/i386/release-notes/ch-whats-new.en.html#id401109
- ^http://neuro.debian.net/pkglists/pkgs-by_release-datasets_%28data%29.html
- ^http://neuro.debian.net/testimonials-topics.html#virtualization
- ^http://neuro.debian.net/derivatives.html
- ^http://www.nitrc.org/plugins/mwiki/index.php/nitrc:NITRC_Computational_Environment
- ^http://wiki.debian.org/UpstreamGuide
- ^http://popcon.debian.org
- ^http://www.nipy.org
- ^http://neuralensemble.org/
References
Fortuna, M. A., Bonachela, J. A., and Levin, S. A. (2011). Evolution of a modular software network. Proc. Natl. Acad. Sci. U.S.A. 108, 19985–19989.
Ghosh, R. (2006). Study on the: Economic Impact of Open Source Software on Innovation and the Competitiveness of the Information and Communication Technologies (ICT) Sector in the EU. Technical Report. European Commission. Available at: http://ec.europa.eu/enterprise/sectors/ict/files/2006-11-20-flossimpact_en.pdf
Gorp, P. V., and Mazanek, S. (2011). SHARE: a web portal for creating and sharing executable research papers. Procedia Comput. Sci. 4, 589–597.
Hanke, M., and Halchenko, Y. O. (2011). Neuroscience runs on GNU/Linux. Front. Neuroinform. 5:8. doi: 10.3389/fninf.2011.00008
Ince, D. C., Hatton, L., and Graham-Cumming, J. (2012). The case for open computer programs. Nature 482, 485–488.
Jackson, I., Schwarz, C., and Debian Project. (2012). Debian Policy Manual [Computer Software Manual], Version 3.9.3.1. Available at: http://www.debian.org/doc/debian-policy
Marcus, D. S., Olsen, T. R., Ramaratnam, M., and Buckner, R. L. (2007). The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics 5, 11–34.
Morin, A., Urban, J., Adams, P. D., Foster, I., Sali, A., Baker, D., and Sliz, P. (2012). Shining light into black boxes. Science 336, 159–160.
Nemoto, K., Dan, I., Rorden, C., Ohnishi, T., Tsuzuki, D., Okamoto, M., Yamashita, F., and Asada, T. (2011). Lin4Neuro: a customized Linux distribution ready for neuroimaging analysis. BMC Med. Imaging 11, 3. doi: 10.1186/1471-2342-11-3
O’Mahony, S., and Ferraro, F. (2007). The emergence of governance in an open source community. Acad. Manage. J. 50, 1079–1106.
Perens, B., Schuessler, E., and Debian Project. (1997). Debian Social Contract [Computer Software Manual], Version 1.2. Available at: http://www.debian.org/social contract
Poline, J.-B., Breeze, J. L., Ghosh, S. S., Gorgolewski, K., Halchenko, Y. O., Hanke, M., Haselgrove, C., Helmer, K. G., Keator, D. B., Marcus, D. S., Poldrack, R. A., Schwartz, Y., Ashburner, J., and Kennedy, D. N. (2012). Data sharing in neuroimaging research. Front. Neuroinformatics 6:9. doi: 10.3389/fninf.2012.000099
Citation: Halchenko YO and Hanke M (2012) Open is not enough. Let’s take the next step: an integrated, community-driven computing platform for neuroscience. Front. Neuroinform. 6:22. doi: 10.3389/fninf.2012.00022
Received: 01 June 2012; Accepted: 05 June 2012;
Published online: 29 June 2012.
Edited by:
Andrew P. Davison, Centre National de la Recherche Scientifique, FranceCopyright: © 2012 Halchenko and Hanke. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: yaroslav.o.halchenko@onerussian.com; michael.hanke@gmail.com
†Yaroslav O. Halchenko and Michael Hanke have contributed equally to this work.