-
PDF
- Split View
-
Views
-
Cite
Cite
Scott V. Dindot, Barbara A. Antalffy, Meenakshi B. Bhattacharjee, Arthur L. Beaudet, The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology, Human Molecular Genetics, Volume 17, Issue 1, 1 January 2008, Pages 111–118, https://doi.org/10.1093/hmg/ddm288
Close - Share Icon Share
Abstract
Loss of function of the maternally inherited allele for the UBE3A ubiquitin ligase gene causes Angelman syndrome (AS), which is characterized by severe neurological impairment and motor dysfunction. In addition, UBE3A lies within chromosome 15q11–q13 region, where maternal, but not paternal, duplications cause autism. The UBE3A gene product, E6-AP, has been shown to function both as an E3 ligase in the ubiquitin proteasome pathway and as a transcriptional coactivator. However, the specific role of E6-AP in the brain, or how loss of function of E6-AP results in AS, is unclear. Herein, we show, using a recombinant transgenic mouse expressing a Ube3aYFP fusion gene, that the maternal Ube3aYFP allele is upregulated and preferentially expressed in neurons, and that the fusion protein, E6-AP:YFP, is enriched in the nucleus and dendrites in vivo. We also show that E6-AP:YFP localizes to the nucleus and to presynaptic and postsynaptic compartments in cultured hippocampal neurons. Furthermore, we show that cerebellar Purkinje cell number and dendritic branching are not affected in Ube3a maternal-deficient mice, but that dendritic spine development, including spine morphology, number and length, is affected on cerebellar Purkinje cells and on pyramidal neurons in the hippocampus and cortex. Collectively, these data suggest that the neurological deficits observed in AS patients and in AS mice may result from specific abnormalities in synaptic development and/or plasticity.
INTRODUCTION
Angelman syndrome (AS) is characterized by severe mental retardation, absence of speech, ataxia and a happy disposition (1,2). The AS gene, UBE3A, encodes the E6-AP ubiquitin ligase and is subject to genomic imprinting, with preferential maternal-specific expression in brain and, more specifically, in neurons but not in glia (3,4). Maternal but not paternal interstitial duplications of chromosome 15q11–q13 cause autism (5). These duplications encompass multiple genes, but UBE3A is a strong candidate to contribute to the autism phenotype in 15q11–q13 duplication cases based on its imprinted status and known causative role in AS (6). Autism spectrum disorders are seen with Prader–Willi syndrome and AS, the two phenotypes associated with paternal and maternal deletions of 15q11–q13, respectively (7,8).
Mice with a maternal null mutation in Ube3a (AS mice) have defects in long-term potentiation (LTP) and manifest motor and behavioral abnormalities that parallel findings in AS in spite of normal cellular architecture in the brain (9). The neurological deficits in AS mice have been directly linked to the postsynaptic calcium/calmodulin kinase type 2 (CaMKII) signaling pathway, as AS mice have increased inhibitory phosphorylation of αCaMKII in the brain (10). Moreover, the learning and behavioral deficits present in AS mice can be rescued through a mutation in CaMKII that prevents its inhibitory phosphorylation (11). Other reports have linked E6-AP to various neurologically relevant proteins including epithelial cell-transforming sequence 2 oncogene (Ect2) (12), neuronal protein interacting specifically with TC10 (nPIST) (13) and myc-binding protein-2 (MYCBP2) (ortholog of Drosophila Highwire) (14), although the functional relevance of these interactions is unclear.
The AS protein, E6-AP, has three isoforms that differ at the N-terminus (15), is a multifunctional E3 ubiquitin ligase and has transcriptional coactivator activity (16); some targets of ubiquitination are known (e.g. p53 oncoprotein), but none has shed light on the AS phenotype or the function of E6-AP in the brain. Although Ube3a is highly expressed in neurons on the basis of in situ hybridization studies, the subcellular localization of E6-AP remains unclear, particularly as it relates the dual transcriptional coactivator and ubiquitin ligase activities, and how its deficiency causes the AS phenotype.
Here, we show, using a knock-in Ube3aYFP fusion reporter mouse, that Ube3aYFP is expressed preferentially from the maternal allele in central neurons but is biallelicly expressed in glial cells, and that the E6-AP:YFP fusion protein localizes both to the nucleus and to the synapse. In addition, we show that mice with maternal deficiency for Ube3a have abnormal dendritic spine morphology and density.
RESULTS
Generation of Ube3aYFP reporter mice
In order to examine the cell-type parental allelic expression of Ube3a and to elucidate the cellular localization of the Ube3a gene product, E6-AP, we produced mice with a knock-in mutation fusing YFP to the C-terminus of E6-AP (Fig. 1A–C; targeted allele, Ube3aYFP; fusion protein, E6-AP:YFP). The Ube3aYFP mice were mated with wild-type (WT) mice to obtain offspring that inherited the Ube3aYFP allele either through the maternal or paternal germline. On the basis of the increased molecular weight of E6-AP:YFP (E6-AP, 100 kDa; E6-AP:YFP, 130 kDa), we used western blotting to distinguish the expression of the Ube3aYFP allele according to parent of origin. Cerebellar lysates from mice inheriting the Ube3aYFP allele maternally, but not paternally, revealed an increase in the molecular weight of E6-AP, indicating preferential maternal expression of the Ube3aYFP allele (Fig. 1D). Maternal-specific expression of Ube3aYFP in cerebellum was further confirmed with an anti-YFP antibody (Fig. 1D). Moreover, the amount of E6-AP:YFP fusion protein detected by western blot was consistent with the levels of endogenous E6-AP protein, indicating that expression of Ube3aYFP was not grossly altered by the generation of the fusion protein (Fig. 1D). In addition, studies with multiple antibodies to E6-AP in WT mice were consistent with the distribution patterns seen with the fusion protein (data not shown).
Generation of Ube3aYFP reporter mice. (A). Schematic illustration of the Ube3a genomic locus (top), Ube3aYFP-targeting construct (TC, middle) and Ube3aYFP-targeted allele (bottom). An SphI restriction site located upstream of the translational stop codon was used to remove the stop codon and to fuse the YFP reporter gene to the C-terminus of E6-AP. (B) Southern blot analysis of wild type (+/+) and Ube3aYFP [E6-AP:YFP (EY) EY/+] positive ES cells. DNA was digested with EcoRV and hybridized with the left arm (LA) probe shown in (A). The fragment sizes for wild type (+/+) and EY (EY/+) alleles are 9.0 and 12.0 kb, respectively. DNA was digested with NdeI and hybridized with the right arm (RA) probe. The fragment sizes for wild type (+/+) and EY (EY/+) alleles are 7.9 and 10.9 kb, respectively. (C) Southern blot analysis of wild type (+/+), Ube3aYFP heterozygous (EY/+) and Ube3aYFP homozygous tail DNA digested with EcoRV and probed with the LA probe. (D) Western blots were performed with wild type (+/+), Ube3aYFP maternal (EY/+) and Ube3aYFP paternal (+/EY) extracts and probed with anti-E6AP (left panel) or anti-YFP (right panel) demonstrating maternal expression of Ube3aYFP in cerebellum. E, EcoRV; N, NdeI; S, SphI.
Analysis of Ube3aYFP allelic expression and cellular localization in vivo
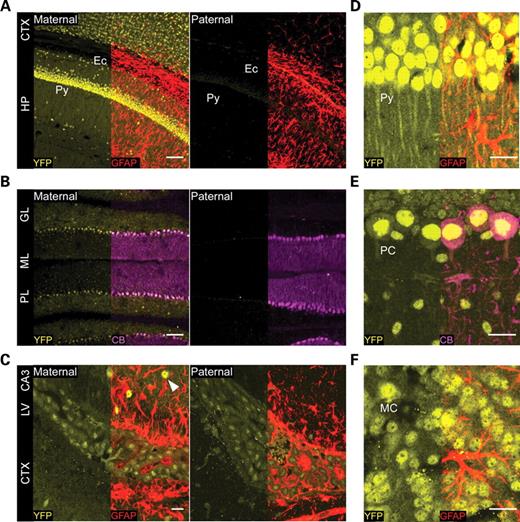
Next, we examined the cell type of the expression of Ube3aYFP according to parent-of-origin, and the subcellular localization of E6-AP:YFP in the brain using confocal imaging. Frozen sections from adult Ube3aYFP maternal and Ube3aYFP paternal brains were immunostained with anti-YFP and anti-GFAP, a glial cell marker, or anti-YFP and anti-calbindin, a cerebellar Purkinje cell marker. Ube3aYFP maternal mice exhibited high levels of expression in the cortical and hippocampal pyramidal neurons (Fig. 2A and D), whereas the expression was only faintly detected in the corresponding regions in Ube3aYFP paternal mice (Fig. 2A). In the cerebellum, Ube3aYFP was expressed in Purkinje cells and in neurons in the granular and molecular layers, although at lower levels (Fig. 2B). Biallelic expression of Ube3aYFP was detected in GFAP-positive cells lining the lateral ventricles, although each allele was expressed at levels lower than seen with the expression of the maternal allele in neurons in the CA3 region of the hippocampus (Fig. 2C). Biallelic expression was limited to GFAP-positive cells lining the ventricles, including the third ventricle (data not shown), and was not readily detectable in GFAP-positive astrocytes in other brain regions. Expression of maternal-derived Ube3aYFP was detected in most brain regions including hippocampus, cortex, thalamus, olfactory bulb and cerebellum. Ube3aYFP paternal expression was only faintly detected in these and other brain regions, aside from the expression detected in GFAP-positive cells lining the ventricles. For each neuronal cell population, E6-AP:YFP localized primarily to the nucleus, but was additionally present in the soma and dendrites as is clearly seen in pyramidal, Purkinje and olfactory bulb neurons (Fig. 2D, E and F). These data indicate that Ube3aYFP undergoes preferential abundant expression of the maternal allele exclusively in neurons, and that E6-AP:YFP localizes to nucleus, cytoplasm of the cell body and processes in these cells.
Ube3aYFP undergoes preferential maternal-specific expression in neurons and biallelic expression in GFAP cells lining the ventricles. (A) Ube3aYFP expression was detected in pyramidal (Py) neurons in the cortex (CTX) and the CA1 region of hippocampus when inherited through the maternal (left), but not paternal (right), germ-line. (B) Ube3aYFP expression was also detected in cerebellar Purkinje cell layer (PL) and in neurons in the granular layer (GR) and molecular layer (ML) when maternally, but not paternally, inherited. (C) Biallelic expression of Ube3aYFP was detected in GFAP-positive cells located adjacent to the lateral ventricles (external capsule cells). Expression of the Ube3aYFP maternal allele was higher in CA3 neurons (white arrow) compared with GFAP-positive cells near the lateral ventricle. Higher magnification image of the CA1 region in the hippocampus (D) and cerebellum (E) demonstrated that maternal-derived Ube3aYFP was highly expressed in neurons. E6-AP:YFP staining was most intense in nuclei but was readily detectable in the cytoplasm of the cell body and in dendrites. In the cerebellum, staining was most intense in Purkinje cell nuclei but was readily detectable in the nuclei of neurons in the molecular layer (ML) and granular layer (GL). Weaker staining was detected in Purkinje cell cytoplasm and processes. (F) Expression of maternal Ube3aEYFP was additionally detected in olfactory neurons. Ec, external capsule; MC, mitral cell; YFP, anti-YFP; GFAP, anti-GFAP; CB, anti-calbindin. Scale bar: (A) and (B) 100 µm; (C), (D), (E) and (F) 20 µm.
E6-AP:YFP localizes to both presynaptic and postsynaptic compartments in dissociated hippocampal neurons in culture
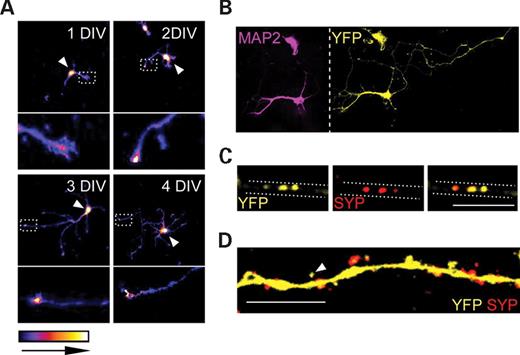
Since E6-AP:YFP was enriched in the nuclei of neurons, and was additionally present in cell body and dendrites, as seen in hippocampal CA1 pyramidal neurons (Fig. 2D), we next analyzed the subcellular localization of E6-AP:YFP in dissociated hippocampal neurons developing in culture derived from mice with a maternally inherited Ube3aYFP allele. Hippocampal neurons at 1–4 days in vitro (DIV) exhibited high levels of E6-AP:YFP in the growth cones of developing neurites and in the nucleus (Fig. 3A). At 4 DIV, anti-MAP2 was used to stain dendrites, and high levels of E6-AP:YFP were detected in the growth cones of both axons and dendrites (Fig. 3B). In order to examine whether E6-AP:YFP was present at the synapse, we analyzed Ube3aYFP neurons cultured for 25 DIV and double-stained with anti-synaptophysin, a presynaptic vesicle marker, and anti-YFP. E6-AP:YFP was highly enriched in the nucleus and was present along axons and dendrites. Images of axons indicated that E6-AP:YFP colocalized with synaptophysin as distinct puncta along axons (Fig. 3C). In dendrites, a different pattern of staining was observed, with E6-AP:YFP being diffusely distributed and not forming distinct puncta. However, E6-AP:YFP was detected in spine-like protrusions along dendritic shafts, some exhibiting positive and negative staining for synaptophysin (Fig. 3D). Collectively, these results indicate that E6-AP:YFP differentially localizes to the nucleus, growth cone and synapse, including presynaptic and postsynaptic compartments, in hippocampal neurons in culture, suggesting that E6-AP may directly regulate development and/or function of synapses.
Intracellular distribution of E6-AP:YFP to the nucleus and synapse in dissociated hippocampal neurons. E6-AP:YFP undergoes differential intracellular trafficking to the growth cone, synapse (presynaptic and postsynaptic) and nucleus in dissociated hippocampal neurons. (A) Confocal images taken of Ube3aYFP maternal-derived neurons at 1, 2, 3 and 4 DIV demonstrated that E6-AP:YFP was enriched in the nucleus (white arrowheads) and growth cones of primary elongating neurites. The ends of primary neurites were magnified to demonstrate enrichment of E6-AP:YFP (dashed boxes are expanded to panels below). (B) Images taken of Ube3aYFP neurons 4 DIV stained with anti-MAP2 and anti-YFP indicated localization of E6-AP:YFP to both axons and dendrites in developing neurons. (C) High magnification confocal images taken of 25 DIV neurons stained with anti-synaptophysin (SYP) and anti-YFP demonstrated distinct clustering of E6-AP:YFP with synaptophysin along axons. (D) Along dendritic shafts, E6-AP:EYFP was diffusely distributed and was present in dendritic spines (white arrowheads). Scale bar: 10 µm in (C) and (D).
Cerebellar Purkinje cell number and dendritic branching are not affected by loss of maternal E6-AP
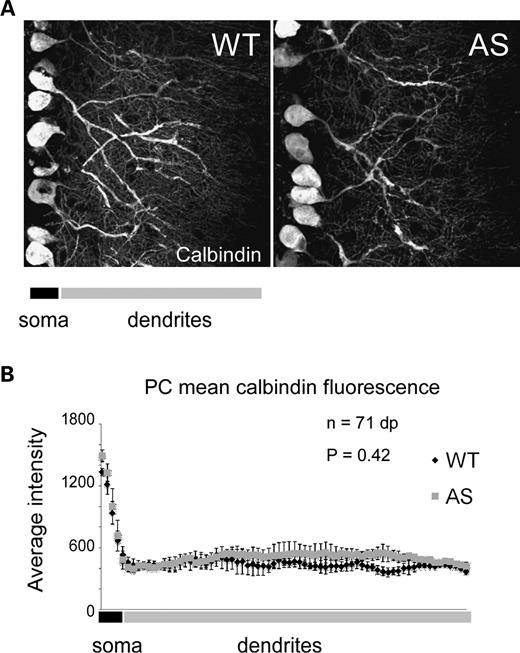
We next evaluated cerebellar Purkinje cell number and dendritic branching in Ube3a maternal-deficient mice (AS mice) (9,17). We analyzed the integrity of cerebellar Purkinje cells in WT and AS mice by examining organization, cell number and dendritic branching through calbindin staining, which immunolabels Purkinje cell soma and dendrites. The distribution and organization of Purkinje cells in AS mice was similar to WT, with no obvious differences being apparent at the light microscope level. Furthermore, we did not detect any differences in Purkinje cell number between WT and AS mice (WT, 11.98 ± 0.83/250 µm PC layer; AS, 11.64 ± 0.19/250 µm; P= 0.7), nor did we detect any differences in calbindin staining of dendritic trees, which is an indirect indicator of Purkinje cell dendritic density and branching (Fig. 4A) (18). These observations are consistent with previous reports (9,19). In other regions in the brain, there were no apparent abnormalities observed by immunohistochemical analysis. Therefore, these data indicate that the neurological deficits observed in AS mice do not arise from abnormalities involving Purkinje cell number or dendritic branching.
Loss of maternal E6-AP does not affect cerebellar Purkinje cell organization or dendritic branching. (A) Representative confocal images showing calbindin staining in WT and AS (maternal null) mice. Purkinje cells in AS mice show similar organization and branching compared with WT. (B) Quantitative confocal analysis of calbindin staining in Purkinje cell dendrites as an indirect indicator of branching. The mean calbindin fluorescence across Purkinje cell soma and dendrites was not statistically significant between WT and AS mice, indicating normal Purkinje cell dendritic branching in AS mice. dp, data points.
AS mice have abnormal dendritic spines on cerebellar Purkinje cells and hippocampal and cortical pyramidal neurons
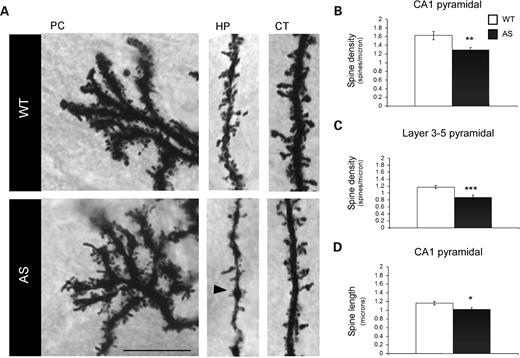
Given the presence of E6-AP:YFP in dendrites in vivo and at synapses in cultured neurons, we performed Golgi staining in WT and AS mice in order to examine dendritic spine morphology and density. Brains from adult WT and AS littermates were extracted and Golgi impregnated. Corresponding sections from WT and AS mice were examined by light microscopy. Images revealed that dendritic spines along Purkinje cells, hippocampal pyramidal and cortical pyramidal neurons in WT mice appeared ‘mushroom like’ with large heads and thin, short necks studded along dendrites. In contrast, spines in AS mice displayed an inconsistent morphology, including high variability in spine neck length and head size. The density of spines also appeared lower in AS mice compared with WT mice (Fig. 5A). Because the high density and crowding of spines in Purkinje cells made detailed analysis difficult, we quantified dendritic spines in hippocampal and cortical pyramidal neurons (Fig. 5A). Secondary apical dendrites close to the cell soma and extending horizontally from pyramidal neurons in the CA1 region were analyzed for dendritic spine density and length in WT and AS mice. Quantification revealed that AS mice showed a significant reduction in spine density compared with WT mice (WT, 1.614 ± 0.076 spines/µm; AS, 1.228 ± 0.055 spines/µm; P= 0.0003; Fig. 5B). A reduction in spines along cortical apical dendrites was also observed in AS mice (WT, 1.172 ± 0.044 spines/µm; AS, 0.882 ± 0.057 spines/µm; P= 0.0009; Fig. 5C). We next quantified spine lengths in hippocampal dendrites, and AS mice exhibited a significant reduction in the overall average spine length compared with WT (WT, 1.16 ± 0.0038 µm; AS, 1.017 ± 0.0454 µm; P= 0.03; Fig. 5D). The pyramidal dendrites in AS mice also exhibited varicosities along the secondary dendritic shaft (Fig. 5A). In addition, many of the dendrites appeared thinner in AS mice relative to WT mice. Collectively, these data indicate that loss of maternal E6-AP results in abnormal dendritic spine development and may underlie the deficits in functional synaptic plasticity observed in AS mice.
Abnormal dendritic spine morphology in mice with maternal deficiency for E6-AP. AS mice exhibited abnormal dendritic spines on cerebellar, cortical and hippocampal neurons, despite normal cellular architecture and dendritic branching. (A) Light micrograph images from Golgi-impregnated Purkinje cell (PC), hippocampal (HP) pyramidal neurons and cortical (CT) pyramidal neurons demonstrated AS mice had reduced dendritic spine density and an abnormal morphology in AS mice compared with WT mice. In addition, AS mice had swellings (black arrowheads) along secondary apical dendrites. (B) and (C) Dendritic spine density was significantly reduced along secondary apical dendrites in CA1 pyramidal neurons and on layer 3–5 cortical pyramidal neurons in AS mice. (D) The average dendritic spine length along apical dendrites in CA1 pyramidal was reduced in AS mice. Scale bar: 10 µm. Error bars are standard error of the mean. *P= 0.03, **P= 0.0003, ***P= 0.0009.
DISCUSSION
In the present study, we examined the cell-type expression and subcellular localization of E6-AP in the brain through a Ube3aYFP knock-in fusion reporter mouse. In addition, we examined the brains of AS mice for yet undetected morphological phenotypes associated with the neurological deficits present in these mice. Our results indicate that Ube3a is highly expressed from the maternal allele specifically in neurons, and that E6-AP differentially localizes to the nucleus and synapse, where it may act locally to regulate synaptic development and plasticity.
Previous studies have indicated that the mouse Ube3a gene undergoes genomic imprinting with preferential expression of the maternal allele in central neurons in the brain (3,4,17). Our results are consistent with these previous observations and demonstrate through in vivo analyses that the paternal allele is expressed at very low-to-undetectable levels in neurons, whereas both alleles are expressed approximately equally in glial cells lining the ventricles. Notably, we observed an upregulation of the maternal allele in neurons relative to the maternal allele expressed in glial cells with biallelic expression. We also observed that E6-AP:YFP was differentially localized between the nucleus and cell body soma, extending to the growth cone and presynaptic and postsynaptic compartments in hippocampal neurons in culture. These findings support a diverse role for E6-AP as a ubiquitin ligase and transcriptional coactivator.
Our observations that AS mice have a normal cellular architecture in the brain but possess dendritic spines that are abnormally shaped and reduced in size and density suggest that E6-AP does not regulate neurogenesis per se, but may function locally to regulate spine development or synaptic plasticity. The presence of E6-AP at the synapse further supports this idea (20), although we cannot exclude a possible contribution of a nuclear role for E6-AP in regulating the structure and/or function of the synapse. Consistent with other mouse models for mental retardation, such as fragile X and Rett syndrome, AS mice clearly possess altered spine morphologies. Fmr1 null mice and fragile X patients have been shown to possess abnormally long dendritic spines as well as increased dendritic spine densities in various neuronal populations (21–27). In contrast, MeCP2-deficient mice and Rett patients have been shown to exhibit reduced dendritic spine densities (28–30). Consistent with our observations in AS mice, Fmr1 and MeCP2 null mice possess apparently normal neuronal maturation and arborization but manifest abnormal synapse development.
Although there have been no specific targets of E6-AP ubiquitination identified to date that would contribute to our understanding of E6-AP synaptic function, there is evidence for effects on the CaMKII pathway. Studies in AS mice have shown that deficiency of Ube3a results in increased inhibitory phosphorylation and reduced postsynaptic density-associated CaMKII (10). Further studies have shown rescue of the AS behavioral phenotypes in CaMKII mutants by introducing an additional mutation at the inhibitory phosphorylation site of αCaMKII; therefore, demonstrating dysregulation of CaMKII is likely the underlying molecular basis for the LTP deficits (11). Consistent with our findings of abnormal dendritic spines in AS mice, CaMKII has been shown to play an important role in both functional and structural synaptic plasticity. Induction of activated CaMKII results in new filopodia growth and spine formation, whereas inhibition of CaMKII activity results in filopodia and spine loss (31). Therefore, loss of E6-AP and the subsequent dysregulation of CaMKII likely lead to impairments in both functional and structural synaptic plasticity.
There is other evidence that is consistent with an important role for E6-AP at the synapse. E6-AP has been shown to interact with MYCBP2, the human ortholog of Drosophila Highwire which is known to regulate synaptic development and function (14,32,33). Given the lack of any apparent organizational or cellular defects in AS mice and the presence of E6-AP at the synapse, we predict that the loss of synaptic E6-AP and the resulting abnormal morphology of dendritic spines in AS mice contribute partially, if not entirely, to the neurological deficits observed in these mice and possibly in AS individuals.
MATERIALS AND METHODS
Generation of Ube3aYFP mice
Genomic DNA from the 129/SvEv mouse strain was used as a template to PCR-amplify an 8.88 kb fragment of the 3′ end of Ube3a including intron 8, exon 9, intron 9, exon 10 and the 3′-UTR (F: 5′-GTTTCGTCACTTCTTTCTTCCGCTC; R: 5′-TGTTAGTTTCCAATGCCTCCTACGC). The PCR fragment was TA-cloned into the TOPO-XL vector (Invitrogen, Carlsbad, CA). The homology arms were digested at an SphI site that is located eight nucleotides upstream of the Ube3a translational stop codon. A linker, which contained the 3′ end of the coding sequence without the translational stop but with an XhoI restriction site, was ligated into the SphI site (F: 5′-CTGTTCTCGAGGAATTCCGCGGTCATG; R: 5′-ACCGCGGAATTCCTCGAGAACAGCATG). An EYFP:loxP:PGK-Neomycin:loxP cassette was ligated into the XhoI site allowing for in-frame fusion of the EYFP gene to the full-length Ube3a. The Ube3aYFP targeting construct was linearized and electroporated into AB2.2 ES cells as described previously (9). G418-resistant clones were screened by Southern blotting using a 5′-PCR probe (F: 5′-GGACTTTGCTTGGTGTTGGT; R: 5′-GGAAGTCAGAAGGGGGAA) and 3′-PCR probe (F: 5′-GGGTCTTTTGAGTTGCGGTA; R: 5′-TCATCTGTCCCAGGGAAAAC). One positive clone was expanded and confirmed for homologous recombination by a second round of Southern blotting using the same 5′- and 3′-probes. The targeted clone was injected into albino C57/BL6 blastocysts by standard procedures. Three low-to-medium percentage chimeras were bred to C57/BL6, and germline transmission of the Ube3aYFP allele was determined by agouti coat color and Southern blot. The Ube3aYFP genotype was maintained on a mixed 129/SvEv and C57/BL6 background.
Western blot analysis
Mice were sacrificed and brains immediately isolated and transferred to ice-cold PBS. Brain tissues were dissected out and immediately transferred to tubes containing ice-cold NP40/SDS buffer (1% Nonidet P-40, 0.01% SDS, 0.1 m Tris–HCl, pH 7.2) and complete Protease Inhibitor Cocktail (Roche Applied Sciences, Indianapolis, IN) and homogenized with a motorized pestle. The lysates were rotated for 15 min at 4°C, centrifuged at 14 000 r.p.m. for 15 min to remove insoluble proteins and then transferred to fresh tubes and put on ice. This was repeated and supernatants were combined into one tube. Protein concentrations were determined using the Bradford reagent (Bio-Rad, Hercules, CA).
Brain tissue homogenates were resolved on a 7.5% SDS–PAGE gel and transferred to a TransBlot nitrocellulose membrane (Bio-Rad). The membranes were blocked in 3% non-fat milk in 0.1% T-TBS (10 mm Tris–HCl, pH 7.5, 150 mm NaCl and 0.1% Tween-20) for 1 h at room temperature. The membranes were incubated in the primary antibody for 1 h at room temperature and then washed three times with T-TBS. The membrane was then incubated in secondary antibody for 1 h at room temperature. The membranes were washed again three times in T-TBS before being developed by enhanced chemiluminescence method (Amersham, Piscataway, NJ). Anti-GFP was diluted at 1:3000 (8371; BD Bioscience, San Jose, CA), and anti-E6-AP (611416; BD Bioscience) was diluted at 1:1000. Secondary antibodies were diluted at 1:3000.
Hippocampal neuronal culture
Primary cultures of hippocampal neurons were obtained from P0-P1 pups of C57/BL6 males crossed to Ube3aYFP females. Hippocampi were dissociated with papain and cultured in a chemically defined medium (Neurobasal medium A; Invitrogen, San Diego, CA) supplemented with B27 (Invitrogen) and penicillin/streptomycin (Invitrogen) on glass coverslips coated with Poly-d-Lysine (Sigma) and collagen (BD Biosciences), as reported previously (34). Cultures were maintained at 37°C in 5% CO2 until use.
Immunostaining
Mice were perfused with ice-cold PBS and 4% paraformaldehyde and tissues harvested as described previously (35). For frozen sections, 45 µm coronal sections were cut on a cryostat and stored in PBS. Sections were washed in PBS and then blocked in T-TBS (10 mm Tris–HCl, pH 7.5, 0.3% TritonX-100) plus 5% normal goat serum for 1–2 h at 4°C. Sections were incubated in primary antibody for 48–72 h at 4°C in a humidified chamber with gentle agitation. Anti-GFP (NB 600–308; Novus Biologicals, Littleton, CO) was used at 1:2000 dilution, anti-GFAP (MAB360; Chemicon, Temecula, CA) at 1:250 and anti-calbindin (C9848; Sigma) at 1:500. Sections were washed three times in T-TBS for 10 min each and then incubated with a fluorescently labeled secondary antibodies (Jackson ImmunoResearch, West Grove, PA) for 24 h at 4°C in the dark. Anti-rabbit Alexa 488 was used at 1:200, and anti-mouse Alexa 557 was used at 1:200 (Invitrogen). Sections were washed again in T-TBS for 20 min each and mounted on glass slides with Vectashield (Vector Laboratories, Burlingame, CA) mounting reagent. Confocal images were obtained using an LSM 510 Zeiss confocal microscope (Zeiss, Oberkochen, Germany).
Immunostaining data analysis
Ube3aYFP parental expression
All images were acquired on a Zeiss LSM 510 confocal microscope with a 10× or 43× oil-immersed objective. The detector and amplifier gain exposure settings were kept the same between sample groups. For anti-GFP fluorescence in Ube3aYFP maternal and Ube3aYFP paternal mice, WT sections were imaged in order to determine the maximum laser intensity and detector gain below the threshold level of fluorescence detection. Ube3aYFP maternal and Ube3aYFP paternal sections were then scanned using these imaging conditions.
Purkinje cell dendritic arborization
For Purkinje cell dendritic arborization, 45 µm cryosections from 6-week-old WT (n= 2) and AS (n= 2) sex-matched littermates were immunostained with anti-calbindin (1:500). All images were acquired on a Zeiss confocal microscope using the 43× objective. The same folia and approximate depth of section were analyzed for each animal. Images were acquired from the Purkinje cell soma extending to the distal end of dendritic branches in the molecular cell layer. Twenty-five sequential, 0.5 µm thick optical images were acquired and were projected using the NIH ImageJ Z-projection function set for average intensity. For each genotype, a 20 × 160 µm rectangular section centered on an individual PC soma was selected from the same region from each animal. The fluorescence intensity profile of this region was calculated using the plot profile in ImageJ. Mean fluorescence (n= 5) for the optical section was calculated every 2.25 µm along PC soma and dendrites. Student’s t-test was performed on the mean fluorescence for the entire optical section (P= 0.421) and on the mean dendritic fluorescence (P= 0.437).
Dendritic spine analysis
Golgi impregnation was performed according to the protocol outlined in the FD Rapid GolgiStain Kit (FD Neurotechnologies, Ellicott, MD). One hundred micrometer serial sections of the cerebellum and hippocampus were cut and mounted. Images were acquired using a Zeiss (Zeiss Imager Z.1) light microscope. For dendritic spine density and length measurements, 30–40 serial z-stacked images spanning the entire dendrite and protruding spines (0.36 µm/image) were acquired using a 100× oil immersion lens. For hippocampal pyramidal neurons, six to seven secondary dendrites branching from the apical dendrite and most proximal to the cell body soma were imaged per animal. For cortical pyramidal neurons, five to six apical dendrites, ∼100 µm from the cell body soma, were imaged per animal. Serial images were analyzed using NIH ImageJ software. Spine density and length were quantified by focusing through each serial image, and counts and measurements were made using the measurement function. For images, 20–25 z-stacked images were projected through deconvolution software. Seven to eight-week-old male and female WT (n= 3) and AS (n= 3) littermate mice were used for the analysis.
Statistics
Microsoft Excel was used for the statistical analysis. For WT and AS comparisons, Student’s t-test was used to determine statistical significance. All data represent mean ± standard error of the mean. A Student’s t-test with a value of P< 0.05 was considered significant (individual P-values are indicated by an asterisk in the figures).
FUNDING
S.V.D and A.L.B. are supported by grants from the National Institutes of Health (F32 HD-049248; R01 HD-037283). This work was supported by Baylor College of Medicine Mental Retardation and Developmental Disabilities Research Center (MRDDRC) NIH grant HD-24064.
ACKNOWLEDGEMENTS
The authors thank Drs Huda Y. Zoghbi, Hugo J. Bellen and Kim Tolias for critical reading of the manuscript.
Conflict of Interest statement. None declared.


![Generation of Ube3aYFP reporter mice. (A). Schematic illustration of the Ube3a genomic locus (top), Ube3aYFP-targeting construct (TC, middle) and Ube3aYFP-targeted allele (bottom). An SphI restriction site located upstream of the translational stop codon was used to remove the stop codon and to fuse the YFP reporter gene to the C-terminus of E6-AP. (B) Southern blot analysis of wild type (+/+) and Ube3aYFP [E6-AP:YFP (EY) EY/+] positive ES cells. DNA was digested with EcoRV and hybridized with the left arm (LA) probe shown in (A). The fragment sizes for wild type (+/+) and EY (EY/+) alleles are 9.0 and 12.0 kb, respectively. DNA was digested with NdeI and hybridized with the right arm (RA) probe. The fragment sizes for wild type (+/+) and EY (EY/+) alleles are 7.9 and 10.9 kb, respectively. (C) Southern blot analysis of wild type (+/+), Ube3aYFP heterozygous (EY/+) and Ube3aYFP homozygous tail DNA digested with EcoRV and probed with the LA probe. (D) Western blots were performed with wild type (+/+), Ube3aYFP maternal (EY/+) and Ube3aYFP paternal (+/EY) extracts and probed with anti-E6AP (left panel) or anti-YFP (right panel) demonstrating maternal expression of Ube3aYFP in cerebellum. E, EcoRV; N, NdeI; S, SphI.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/hmg/17/1/10.1093/hmg/ddm288/2/m_ddm28801.jpeg?Expires=1716413513&Signature=jB2RN2QuT9boayHDATTHk-WKiWZWXbFFiHSPqdkDs6rSHyu0HQyLkM-XYN7jysGQK-rHJttdj3gQ36atj-1bhlSl-8xuzthcTPljN6JOQnRn-Yuz2phGTWEPD-9Iz8W28vEwLmq1O-SWdTVHpfBe8Sogs91KhzXxrJxF9Ua7218YGL7lvuuk6~1N70BzqcSD7M6ceF2Fw9aauCPMu8IzBofTQxi8WC-AKN07~nwsDHhiSqk1noFcvwek5uZMDCZuqBxoHuDCBhMJBQiZgrLFWuOm33OySGi7c2LfnUCd60NtFCwfllFJ2orFAutOZUrWvEGnTWAuNASr6wUACqLkdA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)