-
PDF
- Split View
-
Views
-
Cite
Cite
C. Kaufmann, R. Wehrle, T. C. Wetter, F. Holsboer, D. P. Auer, T. Pollmächer, M. Czisch, Brain activation and hypothalamic functional connectivity during human non-rapid eye movement sleep: an EEG/fMRI study, Brain, Volume 129, Issue 3, March 2006, Pages 655–667, https://doi.org/10.1093/brain/awh686
Close - Share Icon Share
Abstract
Regional differences in sleep EEG dynamics indicate that sleep-related brain activity involves local brain processes with sleep stage specific activity patterns of neuronal populations. Macroscopically, it is not fully understood which cerebral brain regions are involved in the successive discontinuation of wakefulness. We simultaneously used EEG and functional MRI on 9 subjects (6 female: mean = 24.1 years, 3 male: mean = 26.0 years) and analyzed local blood oxygenation level dependent signal changes linked to the transition from wakefulness to different non-rapid eye movement (NREM) sleep stages (according to Rechtschaffen and Kales) of the first sleep cycles after 36 h of total sleep deprivation. Several brain regions throughout the cortex, the limbic lobe, the thalamus, the caudate nucleus, as well as midbrain structures, such as the mammillary body/hypothalamus, showed reduced activity during NREM sleep across all sleep stages. Additionally, we found deactivation patterns specific to NREM sleep stages compared with wakefulness suggesting that a synchronized sleeping state can be established only if these regions interact in a well-balanced way. Sleep stage 2, which is usually linked to the loss of self-conscious awareness, is associated with signal decreases comprising thalamic and hypothalamic regions, the cingulate cortex, the right insula and adjacent regions of the temporal lobe, the inferior parietal lobule and the inferior/middle frontal gyri. The hypothalamic region known to be of particular importance in the regulation of the sleep–wake cycle shows specific temporally correlated network activity with the cortex while the system is in the sleeping state, but not during wakefulness. We describe a specific pattern of decreased brain activity during sleep and suggest that this pattern must be synchronized for establishing and maintaining sleep.
Introduction
When von Economo discussed ‘sleep as a problem of localisation’, he related his previous description of encephalitic lethargica to a ‘grey substance spreading over the posterior and lateral walls of the third ventricle and reaching laterally also into the hypothalamus nuclei’ (von Economo, 1930). He claimed an anatomically relatively diffuse ‘sleep centre’ responsible for sleep regulation. We are now more precisely aware of this sleep centre, which may be divided into the sleep-promoting ventrolateral preoptic nucleus (VLPO) and the wake-promoting posterior lateral hypothalamus (Saper et al., 2001). Although consciousness abruptly ceases when falling asleep, the activity of the cerebral cortex which is a sine qua non for conscious experience (Creutzfeld, 1995; Muzur et al., 2002) does not fade away across all sleep stages. Since the discovery of awake-like electroencephalography (EEG) activity during rapid eye movement (REM) sleep, we have been aware that the cerebral cortex is not passively on standby while we are asleep. Accordingly, the electrophysiological description of human sleep (Dement and Kleitman, 1957; Rechtschaffen and Kales, 1968) reveals distinct patterns of repetitious cerebral activity across the sleeping period.
Regional differences in sleep EEG dynamics suggest that these changes in activity are not a global phenomenon but rather involve local brain processes with a different regional involvement of neuronal populations (Werth et al., 1997; Finelli et al., 2001; Ferrara et al., 2002). The current gold standard criteria for the electrophysiological characterization of sleep are based on 30 s temporal resolution data (Rechtschaffen and Kales, 1968) but ignore spatial information about distinct cerebral activity while asleep (Himanen and Hasan, 2000). Diagnostics of several sleep disturbances rely heavily on the sleep stage criteria once suggested. Insomnia illustrates our lack of knowledge concerning the functional significance of these criteria as there is often a remarkable dissociation between the subjective experience of sleep disruptions and the associated EEG parameters (Drummond et al., 2004; Nofzinger, 2005).
Several studies have been carried out combining EEG and imaging methods in order to explore regional specific brain activity during sleep (Buchsbaum et al., 1989; Maquet et al., 1990, 1992, 1996, 1997; Madsen et al., 1991a, b, c; Braun et al., 1997, 1998; Hofle et al., 1997; Nofzinger et al., 1997, 2002; Andersson et al., 1998; Kajimura et al., 1999; Løvblad et al., 1999; Finelli et al., 2000; Peigneux et al., 2001; Balkin et al., 2002; Kjaer et al., 2002). Almost all of these studies used positron emission tomography (PET) or single photon emission computed tomography (SPECT) with the exception of Løvblad et al. who used functional MRI (fMRI). The findings obtained from these studies suggest a global decrease in cerebral and thalamic activity during non-rapid eye movement (NREM) sleep, which is in accordance with the slow potential activity measured with EEG. More specifically, light sleep seems to be characterized by decreased activity in the frontal and parietal cortices, and in the thalamus. As compared with wakefulness the decline of activity is continued during slow-wave sleep (SWS) with an additional decrease in activity within the basal ganglia. On the contrary, REM sleep is accompanied by increased metabolism within the pons, the limbic system and the occipital cortex (secondary visual cortex) and diminished metabolism within parietal and prefrontal regions. Both REM and NREM sleep show diminished brain activity in prefrontal and parietal regions compared with wakefulness. The findings for several other brain regions, as for cerebellar and medial temporal areas, hypothalamic nuclei and paralimbic regions such as the cingulate cortex are still not conclusive.
In general, PET studies show a relatively low spatial as well as temporal resolution compared with fMRI, which can overcome these limitations allowing for a more precise detection of brain activity during sleep. We recently reported sleep stage specific blood oxygenation level dependent (BOLD) signal decreases associated with transient increases in EEG hyperpolarization upon acoustic stimulation (Czisch et al., 2002, 2004). However, processing of sensory information interferes with a putatively dynamic pattern of brain activity during the sleep–wake cycle. Periods free of stimulation would be needed to allow for mapping of brain activations associated with sleep stages. In the present study, we therefore compared the baseline activity across sleep stages by relating local BOLD signal changes to the sleep stage classifications derived from simultaneous polysomnographic recordings without applying external stimulation. Furthermore, as only fMRI allows for a comparison of activity across the brain, and identification of brain regions which alter their activity synchronously with other areas, we demonstrate sleep specific interaction of the hypothalamus, which is known to be of central importance for the regulation of the sleep–wake cycle, with other brain regions by calculating functional connectivity on the basis of the hypothalamic time curves.
Methods
Subjects
Fourteen young healthy paid volunteers gave written informed consent according to the institutional guidelines before participating in this study, which was approved by the local Ethical Committee. The subjects had no history of neurological and psychiatric disorders, or substance abuse, and had no sleep disturbances or recent time zone shifts. Five subjects were deaf, and the initial idea that deaf persons might be less influenced by the scanner noise and thus be better able to fall asleep has proven wrong as all of our subjects reported raised sensitivity towards the gradient switches (resulting in vibrations) during data acquisition. We included these subjects in the analysis since their sleep stage scores did not differ from the hearing subjects (see Results). In total, data from five subjects had to be discarded because they were not able to fall asleep in the uncomfortable and noisy laboratory environment (two subjects), because of movement artefacts, because they immediately fell asleep and thus did not provide baseline data for the awake state or because of technical issues not related to the subject. Data from 9 out of 14 subjects—four of them being deaf—were finally included in the analysis. Ages ranged from 21.9 to 29.3 years with a mean of 24.7 years (6 female: M = 24.1 years, 3 male: M = 26.0 years).
Sleep deprivation procedure and instructions
All subjects underwent a habituation fMRI session. Prior to the second session, which took place within a week, subjects underwent total sleep deprivation for 1 day, i.e. 36 h, to increase the tendency to fall asleep despite the restraining experimental setting within the scanner. Sleep deprivation was controlled using a wrist actigraph. MRI experiments started between 10 and 11 p.m. All subjects wore ear muffs as well as a headphone-like ear protection, and each subject's head was carefully immobilized with a vacuum cushion to minimize movement artefacts. The scanning room was completely darkened during the experiment. Subjects were laid as comfortably as possible backward onto the scanner ‘bed’. They were instructed to stay awake for ∼5–10 min with eyes closed after the scan was started, and were then allowed to fall asleep. The scans were repeated up to three times, if possible. The session was stopped when the volunteer was either completely awake or started to feel uncomfortable, with a typical session lasting between 1 and 3 h.
Simultaneous fMRI and EEG experiments
fMRI data acquisition and analysis
Imaging was performed on a 1.5 tesla scanner (Signa Echospeed, General Electric, Milwaukee, USA) using a standard GE imaging headcoil. Functional T2*-weighted images with a matrix size of 128 × 128 (FOV 28 × 28 cm2, nominal voxel dimensions: 2.1875 × 2.1875 × 5 mm3) were obtained with an echoplanar single shot pulse sequence (EPI) using an axial slice orientation. Repetition time (TR) was 10 s, flip angle 90° and echo time (TE) 60 ms. The volume acquired covered 20 slices. The first 5 of the 200 acquired images were excluded from further analysis to avoid non steady-state effects due to T1 saturation. Scanning time therefore was 32 min 30 s. We had to choose a TR of 10 s because of technical constraints. Image processing was carried out using statistical parametric mapping (SPM99) and statistical analysis with SPM2 were used (Friston et al., 1995). After defining the anterior and posterior commissural line all volumes were realigned to the first volume. Datasets with more than 2 mm motion in any direction were excluded from further analysis. The estimated translational movement parameters were ∼0.5–1.5 mm. The mean image (built on the basis of all realigned volumes) was spatially normalized into standard stereotactic space using an EPI template (SPM99 standard template from the Montreal Neurological Institute). We estimated the Talairach coordinates from the subsequently derived SPM maps with a non-linear transform of MNI to Talairach (different linear transforms to different brain regions) (Brett et al., 2002). Next, we estimated global effects from the images using a voxel-level linear model of the global signal (Macey et al., 2004) to remove effects of signal drift not related to the conditions. Effects that match the global signal are removed from the voxel's time course based on the assumption that the global signal is replicated in the same pattern throughout the brain with varying magnitudes. The data were then smoothed using a full-width at half maximum isotropic Gaussian kernel of 8 mm. Data analysis was performed by modelling the different conditions (sleep stages) as stimulus functions with the movement parameters as regressors of no interest within the context of the general linear model. Applying a multi-level approach we accounted for intra-subject variance in a fixed effects analysis, and for between-subject variance in a random effects analysis (subject by response interaction). As we had nine subjects left for the random effects analysis we chose a less conservative threshold for an alpha level of uncorrected 0.001 (k = 25). An extent threshold of 25 voxels was chosen in order to not include clusters which are considerably smaller than the estimated resolution after the image post-processing steps. Although the thresholded results for some contrasts (i.e. Wakefulness versus Sleep, Wakefulness more than S1, Wakefulness more than S2, Awake more than SWS and Hypothalamic connectivity) survived P values corrected for whole brain volume, we present all results equally thresholded to standardize comparisons. We applied several contrasts for each condition and one for the main condition effect where the sleep stages were tested against the awake ‘rest’ condition. From the resulting SPM maps of the condition effect, after smoothing the data with 4 mm full-width at half maximum to enhance sensitivity for subcortical structures, we chose a region of interest in the hypothalamus according to the definitions of the Talairach coordinate system (5 × 3 × 10 mm) (Lancaster et al., 2000). Within these ROIs we extracted the averaged hypothalamic time series and modelled them as regressors of interest in a second SPM analysis to determine functional connectivity between the awake and sleeping conditions. The localization of the results is presented according to the Talairach Daemon (Lancaster et al., 2000) as integrated in mri3dX (Version 5.6) (Singh, 2004; http://www.aston.ac.uk/lhs/staff/singhkd/mri3dX).
EEG data and analysis
Polysomnographic recordings were performed using a MR-compatible EEG system (Schwarzer, Munich, Germany) according to the international ten-twenty electrode system with eight channels (F3, F4, C3, C4, P3, P4, O1 and O2 versus common average reference), an electrooculogram, a chin electromyogram and a three-lead electrocardiogram. The sampling rate was 500 Hz, and band width was set to 0.5–70 Hz for EEG. All necessary precautions were taken to guarantee the safe recording of the electrophysiological signals during image acquisition, and careful electrode placement could sufficiently suppress cardioballistic artefacts in any of the recording channels. EEG post-processing using a Fourier filtering algorithm (Hoffmann et al., 2000) allowed elimination of scanner artefacts. Briefly, the algorithm compares the power spectrum observed during MR imaging and the combined spectrum of three referential 10 s epochs representing artefact-free EEG of different vigilance states during the same session. All frequency bands (resolution 0.1 Hz) in the EEG power spectrum obtained during scans exceeding more than twice the power of the unaffected reference spectrum were removed. This correction results in sufficient suppression of scanner-induced artefacts with only a minor reduction of the original frequencies while phases are correctly retained. Since the frequency components of the gradient-induced artefacts solely depend on the timing of the imaging experiment and because their amplitude is much larger than subject-specific influences, the correction algorithm removes identical frequency components in all subjects and conditions. The complete recording including fMRI periods was then evaluated off-line (Czisch et al., 2002, 2004) to determine sleep stages in 30 s epochs.
Results
Sleep stage scoring according to Rechtschaffen and Kales
We obtained 15 fMRI/EEG scans from nine subjects (two scans from six subjects, four of them were female and two male). According to Rechtschaffen and Kales' sleep stage criteria all subjects reached sleep stage 2 (S2), seven subjects sleep stage 3 (S3) and four subjects sleep stage 4 (S4) within the scanning duration of 32 min 30 s. Altogether the subjects spent 102 min awake, 88.2 min in S1, 192.8 min in S2, 46.8 min in S3 and 57.3 min in S4 (seeTable 1). The deaf subjects did not show significantly altered sleep stage distribution as compared with the hearing subjects [F(4,10) = 1.074, P = 0.419].
Duration of NREM sleep stages of nine subjects in minutes (of 32 min 30 s which was the duration of one scan)
| Minutes . | Awake . | S1 . | S . | SWS . | ||||
|---|---|---|---|---|---|---|---|---|
| Sleep stage scoring according to 30 s epochs | ||||||||
| Mean | 6.8 | 5.9 | 12.9 | 6.9 | ||||
| SD | 4.7 | 2.9 | 4.5 | 3.9 | ||||
| Sum | 102.3 | 88.2 | 192.8 | 104.1 | ||||
| % Sum | 21 | 18 | 39.5 | 21.5 | ||||
| Minutes . | Awake . | S1 . | S . | SWS . | ||||
|---|---|---|---|---|---|---|---|---|
| Sleep stage scoring according to 30 s epochs | ||||||||
| Mean | 6.8 | 5.9 | 12.9 | 6.9 | ||||
| SD | 4.7 | 2.9 | 4.5 | 3.9 | ||||
| Sum | 102.3 | 88.2 | 192.8 | 104.1 | ||||
| % Sum | 21 | 18 | 39.5 | 21.5 | ||||
The values denote the duration when applying the Rechtschaffen and Kales criteria.
Duration of NREM sleep stages of nine subjects in minutes (of 32 min 30 s which was the duration of one scan)
| Minutes . | Awake . | S1 . | S . | SWS . | ||||
|---|---|---|---|---|---|---|---|---|
| Sleep stage scoring according to 30 s epochs | ||||||||
| Mean | 6.8 | 5.9 | 12.9 | 6.9 | ||||
| SD | 4.7 | 2.9 | 4.5 | 3.9 | ||||
| Sum | 102.3 | 88.2 | 192.8 | 104.1 | ||||
| % Sum | 21 | 18 | 39.5 | 21.5 | ||||
| Minutes . | Awake . | S1 . | S . | SWS . | ||||
|---|---|---|---|---|---|---|---|---|
| Sleep stage scoring according to 30 s epochs | ||||||||
| Mean | 6.8 | 5.9 | 12.9 | 6.9 | ||||
| SD | 4.7 | 2.9 | 4.5 | 3.9 | ||||
| Sum | 102.3 | 88.2 | 192.8 | 104.1 | ||||
| % Sum | 21 | 18 | 39.5 | 21.5 | ||||
The values denote the duration when applying the Rechtschaffen and Kales criteria.
SPM group analysis
All fMRI results reported relate to random effects analysis. We combined S3 and S4 into SWS because not all subjects reached S4, and because S3/S4 are, by definition, very similar according to their EEG reflecting high delta activity.
Comparing BOLD-related brain activity of NREM sleep stages with the resting awake state
In several brain regions activity was reduced during all NREM sleep stages. First, most of these deactivated areas are located in the frontal lobes [inferior frontal gyrus (IFG), middle frontal gyrus (MFG), superior frontal gyrus (SFG), medial frontal gyrus (MedFG), precentral gyrus and paracentral lobule] with a predominance in the right cerebrum (the laterality index for the number of activated voxels is 17). Secondly, regions of the limbic lobe such as the anterior cingulate cortex (ACC) and PCG were also less activated during NREM sleep. Furthermore, the anterior nucleus of the thalamus and the body of the caudate nucleus showed reduced activity, again with a predominance in the right hemisphere. Thirdly, temporal [superior temporal gyrus (STG)], parietal [post-central gyrus, inferior parietal lobule (IPL)], occipital (cuneus, precuneus) as well as insular activation [restricted to the right anterior Brodmann area (BA) 13] diminished during all NREM sleep stages. Finally, midbrain structures such as the mammillary body/hypothalamus reduced their activity while asleep.
BOLD activity during wakefulness more than during stage 1
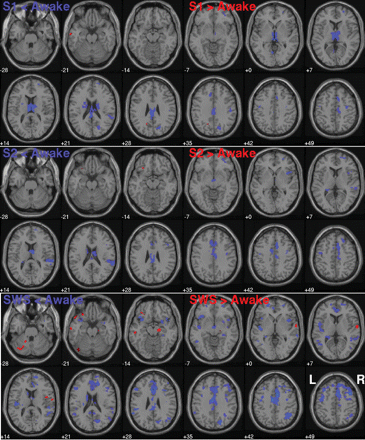
During S1 deactivations in thalamic and cingulate structures were most prominent. Specifically, we found mostly bilaterally less activity during S1 in limbic structures (PCG, dorsal part of the cingulate gyrus, thalamus and caudate nucleus), the frontal lobes (MedFG, MFG, precentral gyrus and SFG), the occipital lobes (precuneus, cuneus and lingual gyrus) and the insula, and less pronounced activity in the IPL and temporal lobes (Table 2 and Fig. 1).
Transverse slice view: The coloured areas represent BOLD-related activations and are superimposed onto a T1-weighted MRI of a male individual as supplied with SPM2. Blue denotes less activation during the sleep stages and red more activation during sleep stages. The first row shows the comparison ‘Awake versus Sleep stage 1’, the second ‘Awake versus Sleep stage 2’ and the third row ‘Awake versus SWS’ (P = 0.001, k = 25, not corrected).
Location of activated voxels in stereotactic space (MNI: x y z) for the comparison of Awake versus Sleep stage 1 (S1)
| Anatomical region (Brodmann area) . | Hemisphere . | x . | y . | z . | No. voxels . | Z-value . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Awake more than S1 | ||||||||||||
| Posterior cingulate gyrus (23) | L | −3 | −35 | 23 | 232 | 4.73 | ||||||
| L | −1 | −35 | 20 | 886 | 4.68 | |||||||
| Cingulate gyrus (24) | L | −11 | 24 | 37 | 818 | 4.46 | ||||||
| 0 | −29 | 31 | 120 | 4.25 | ||||||||
| R | 9 | 13 | 46 | 516 | 4.2 | |||||||
| MedFG (10) | L | −11 | 25 | 37 | 81 | 4.46 | ||||||
| R | 9 | 13 | 47 | 180 | 4.2 | |||||||
| Precuneus | R | 21 | −73 | 25 | 468 | 4.42 | ||||||
| Thalamus | L | −5 | −15 | 15 | 1534 | 4.38 | ||||||
| Pulvinar | R | 3 | −23 | 5 | 700 | 4.25 | ||||||
| R | 5 | −7 | 13 | 1744 | 4.21 | |||||||
| Medial dorsal nucleus | 0 | −11 | 3 | 18 | 3.79 | |||||||
| Precuneus | R | 21 | −69 | 24 | 304 | 4.38 | ||||||
| Cuneus | L | −5 | −65 | 3 | 56 | 4.31 | ||||||
| R | 13 | −71 | 32 | 60 | 3.97 | |||||||
| Lingual gyrus | L | −5 | −65 | 2 | 54 | 4.31 | ||||||
| Caudate | R | 15 | −1 | 19 | 48 | 4.12 | ||||||
| Precentral gyrus | R | 39 | −11 | 57 | 276 | 4.1 | ||||||
| L | −45 | −9 | 45 | 24 | 3.66 | |||||||
| MFG (6, 9, 10) | R | 33 | −11 | 49 | 172 | 4.05 | ||||||
| Putamen | L | −23 | −1 | 18 | 22 | 4.05 | ||||||
| Paracentral lobule | R | 13 | −25 | 51 | 44 | 3.73 | ||||||
| STG (42) | R | 61 | −41 | 20 | 24 | 3.71 | ||||||
| R | 23 | 59 | 13 | 72 | 3.69 | |||||||
| SFG (10, 38) | L | −7 | 5 | 57 | 5 | 3.53 | ||||||
| Claustrum | L | −27 | −3 | 18 | 10 | 3.69 | ||||||
| Post-central gyrus | R | 51 | −19 | 34 | 20 | 3.68 | ||||||
| Insula (13) | R | 53 | −37 | 20 | 8 | 3.68 | ||||||
| IPL (40) | R | 61 | −41 | 23 | 12 | 3.6 | ||||||
| S1 more than Awake | ||||||||||||
| Precuneus | L | −17 | −59 | 31 | 16 | 4.1 | ||||||
| Middle temporal gyrus | L | −65 | −9 | −17 | 6 | 3.97 | ||||||
| Anatomical region (Brodmann area) . | Hemisphere . | x . | y . | z . | No. voxels . | Z-value . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Awake more than S1 | ||||||||||||
| Posterior cingulate gyrus (23) | L | −3 | −35 | 23 | 232 | 4.73 | ||||||
| L | −1 | −35 | 20 | 886 | 4.68 | |||||||
| Cingulate gyrus (24) | L | −11 | 24 | 37 | 818 | 4.46 | ||||||
| 0 | −29 | 31 | 120 | 4.25 | ||||||||
| R | 9 | 13 | 46 | 516 | 4.2 | |||||||
| MedFG (10) | L | −11 | 25 | 37 | 81 | 4.46 | ||||||
| R | 9 | 13 | 47 | 180 | 4.2 | |||||||
| Precuneus | R | 21 | −73 | 25 | 468 | 4.42 | ||||||
| Thalamus | L | −5 | −15 | 15 | 1534 | 4.38 | ||||||
| Pulvinar | R | 3 | −23 | 5 | 700 | 4.25 | ||||||
| R | 5 | −7 | 13 | 1744 | 4.21 | |||||||
| Medial dorsal nucleus | 0 | −11 | 3 | 18 | 3.79 | |||||||
| Precuneus | R | 21 | −69 | 24 | 304 | 4.38 | ||||||
| Cuneus | L | −5 | −65 | 3 | 56 | 4.31 | ||||||
| R | 13 | −71 | 32 | 60 | 3.97 | |||||||
| Lingual gyrus | L | −5 | −65 | 2 | 54 | 4.31 | ||||||
| Caudate | R | 15 | −1 | 19 | 48 | 4.12 | ||||||
| Precentral gyrus | R | 39 | −11 | 57 | 276 | 4.1 | ||||||
| L | −45 | −9 | 45 | 24 | 3.66 | |||||||
| MFG (6, 9, 10) | R | 33 | −11 | 49 | 172 | 4.05 | ||||||
| Putamen | L | −23 | −1 | 18 | 22 | 4.05 | ||||||
| Paracentral lobule | R | 13 | −25 | 51 | 44 | 3.73 | ||||||
| STG (42) | R | 61 | −41 | 20 | 24 | 3.71 | ||||||
| R | 23 | 59 | 13 | 72 | 3.69 | |||||||
| SFG (10, 38) | L | −7 | 5 | 57 | 5 | 3.53 | ||||||
| Claustrum | L | −27 | −3 | 18 | 10 | 3.69 | ||||||
| Post-central gyrus | R | 51 | −19 | 34 | 20 | 3.68 | ||||||
| Insula (13) | R | 53 | −37 | 20 | 8 | 3.68 | ||||||
| IPL (40) | R | 61 | −41 | 23 | 12 | 3.6 | ||||||
| S1 more than Awake | ||||||||||||
| Precuneus | L | −17 | −59 | 31 | 16 | 4.1 | ||||||
| Middle temporal gyrus | L | −65 | −9 | −17 | 6 | 3.97 | ||||||
Location of activated voxels in stereotactic space (MNI: x y z) for the comparison of Awake versus Sleep stage 1 (S1)
| Anatomical region (Brodmann area) . | Hemisphere . | x . | y . | z . | No. voxels . | Z-value . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Awake more than S1 | ||||||||||||
| Posterior cingulate gyrus (23) | L | −3 | −35 | 23 | 232 | 4.73 | ||||||
| L | −1 | −35 | 20 | 886 | 4.68 | |||||||
| Cingulate gyrus (24) | L | −11 | 24 | 37 | 818 | 4.46 | ||||||
| 0 | −29 | 31 | 120 | 4.25 | ||||||||
| R | 9 | 13 | 46 | 516 | 4.2 | |||||||
| MedFG (10) | L | −11 | 25 | 37 | 81 | 4.46 | ||||||
| R | 9 | 13 | 47 | 180 | 4.2 | |||||||
| Precuneus | R | 21 | −73 | 25 | 468 | 4.42 | ||||||
| Thalamus | L | −5 | −15 | 15 | 1534 | 4.38 | ||||||
| Pulvinar | R | 3 | −23 | 5 | 700 | 4.25 | ||||||
| R | 5 | −7 | 13 | 1744 | 4.21 | |||||||
| Medial dorsal nucleus | 0 | −11 | 3 | 18 | 3.79 | |||||||
| Precuneus | R | 21 | −69 | 24 | 304 | 4.38 | ||||||
| Cuneus | L | −5 | −65 | 3 | 56 | 4.31 | ||||||
| R | 13 | −71 | 32 | 60 | 3.97 | |||||||
| Lingual gyrus | L | −5 | −65 | 2 | 54 | 4.31 | ||||||
| Caudate | R | 15 | −1 | 19 | 48 | 4.12 | ||||||
| Precentral gyrus | R | 39 | −11 | 57 | 276 | 4.1 | ||||||
| L | −45 | −9 | 45 | 24 | 3.66 | |||||||
| MFG (6, 9, 10) | R | 33 | −11 | 49 | 172 | 4.05 | ||||||
| Putamen | L | −23 | −1 | 18 | 22 | 4.05 | ||||||
| Paracentral lobule | R | 13 | −25 | 51 | 44 | 3.73 | ||||||
| STG (42) | R | 61 | −41 | 20 | 24 | 3.71 | ||||||
| R | 23 | 59 | 13 | 72 | 3.69 | |||||||
| SFG (10, 38) | L | −7 | 5 | 57 | 5 | 3.53 | ||||||
| Claustrum | L | −27 | −3 | 18 | 10 | 3.69 | ||||||
| Post-central gyrus | R | 51 | −19 | 34 | 20 | 3.68 | ||||||
| Insula (13) | R | 53 | −37 | 20 | 8 | 3.68 | ||||||
| IPL (40) | R | 61 | −41 | 23 | 12 | 3.6 | ||||||
| S1 more than Awake | ||||||||||||
| Precuneus | L | −17 | −59 | 31 | 16 | 4.1 | ||||||
| Middle temporal gyrus | L | −65 | −9 | −17 | 6 | 3.97 | ||||||
| Anatomical region (Brodmann area) . | Hemisphere . | x . | y . | z . | No. voxels . | Z-value . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Awake more than S1 | ||||||||||||
| Posterior cingulate gyrus (23) | L | −3 | −35 | 23 | 232 | 4.73 | ||||||
| L | −1 | −35 | 20 | 886 | 4.68 | |||||||
| Cingulate gyrus (24) | L | −11 | 24 | 37 | 818 | 4.46 | ||||||
| 0 | −29 | 31 | 120 | 4.25 | ||||||||
| R | 9 | 13 | 46 | 516 | 4.2 | |||||||
| MedFG (10) | L | −11 | 25 | 37 | 81 | 4.46 | ||||||
| R | 9 | 13 | 47 | 180 | 4.2 | |||||||
| Precuneus | R | 21 | −73 | 25 | 468 | 4.42 | ||||||
| Thalamus | L | −5 | −15 | 15 | 1534 | 4.38 | ||||||
| Pulvinar | R | 3 | −23 | 5 | 700 | 4.25 | ||||||
| R | 5 | −7 | 13 | 1744 | 4.21 | |||||||
| Medial dorsal nucleus | 0 | −11 | 3 | 18 | 3.79 | |||||||
| Precuneus | R | 21 | −69 | 24 | 304 | 4.38 | ||||||
| Cuneus | L | −5 | −65 | 3 | 56 | 4.31 | ||||||
| R | 13 | −71 | 32 | 60 | 3.97 | |||||||
| Lingual gyrus | L | −5 | −65 | 2 | 54 | 4.31 | ||||||
| Caudate | R | 15 | −1 | 19 | 48 | 4.12 | ||||||
| Precentral gyrus | R | 39 | −11 | 57 | 276 | 4.1 | ||||||
| L | −45 | −9 | 45 | 24 | 3.66 | |||||||
| MFG (6, 9, 10) | R | 33 | −11 | 49 | 172 | 4.05 | ||||||
| Putamen | L | −23 | −1 | 18 | 22 | 4.05 | ||||||
| Paracentral lobule | R | 13 | −25 | 51 | 44 | 3.73 | ||||||
| STG (42) | R | 61 | −41 | 20 | 24 | 3.71 | ||||||
| R | 23 | 59 | 13 | 72 | 3.69 | |||||||
| SFG (10, 38) | L | −7 | 5 | 57 | 5 | 3.53 | ||||||
| Claustrum | L | −27 | −3 | 18 | 10 | 3.69 | ||||||
| Post-central gyrus | R | 51 | −19 | 34 | 20 | 3.68 | ||||||
| Insula (13) | R | 53 | −37 | 20 | 8 | 3.68 | ||||||
| IPL (40) | R | 61 | −41 | 23 | 12 | 3.6 | ||||||
| S1 more than Awake | ||||||||||||
| Precuneus | L | −17 | −59 | 31 | 16 | 4.1 | ||||||
| Middle temporal gyrus | L | −65 | −9 | −17 | 6 | 3.97 | ||||||
BOLD activity during wakefulness more than during stage 2
During S2 deactivations in cingulate (as well as medial and superior frontal) and superior temporal structures were most prominent, followed by thalamic/hypothalamic decreases in activation. More precisely, deactivations were related to regions in the temporal lobes [STG, middle temporal gyrus (MTG)] with a dominance of the right hemisphere, the right parietal lobe (IPL), the limbic lobe (cingulate gyrus, thalamus and hypothalamus), the frontal lobes (MedFG, SFG and right IFG) and the insula in the right hemisphere (Table 3 and Fig. 1).
Location of activated voxels in stereotactic space (MNI: x y z) for the comparison of Awake versus Sleep stage 2 (S2)
| Anatomical region (Brodmann area) . | Hemisphere . | x . | y . | z . | No. voxels . | Z-value . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Awake more than S2 | ||||||||||||
| STG (42) | R | 61 | −41 | 20 | 1404 | 4.85 | ||||||
| Cingulate gyrus (24, 32) | R | 9 | 9 | 46 | 3476 | 4.66 | ||||||
| L | −9 | 3 | 45 | 2404 | 4.2 | |||||||
| 0 | −29 | 31 | 116 | 3.92 | ||||||||
| MedFG | R | 9 | 9 | 47 | 1084 | 4.66 | ||||||
| L | −7 | 7 | 47 | 146 | 4.05 | |||||||
| Thalamus, anterior nucleus | R | 7 | −7 | 17 | 368 | 4.45 | ||||||
| R | 7 | −9 | 17 | 236 | 4.41 | |||||||
| L | −5 | −3 | 14 | 2 | 3.29 | |||||||
| Mammillary body/hypothalamus | L | −5 | −9 | −7 | 406 | 4.11 | ||||||
| Precentral gyrus | R | 37 | −7 | 41 | 230 | 4.36 | ||||||
| L | −47 | −7 | 49 | 288 | 3.54 | |||||||
| IPL (40) | R | 61 | −41 | 23 | 508 | 4.24 | ||||||
| SFG | R | 27 | 55 | −5 | 1024 | 4.2 | ||||||
| MFG | R | 27 | 55 | −6 | 608 | 4.2 | ||||||
| Insula | R | 39 | 13 | 3 | 792 | 4.05 | ||||||
| Post-central gyrus | R | 67 | −29 | 17 | 188 | 4.03 | ||||||
| Caudate | L | −23 | −37 | 9 | 87 | 3.92 | ||||||
| R | 5 | 1 | 14 | 78 | 3.77 | |||||||
| IFG (45) | R | 47 | 17 | 5 | 186 | 3.79 | ||||||
| Paracentral lobule | R | 7 | −19 | 45 | 196 | 3.63 | ||||||
| Transverse temporal gyrus | R | 51 | −27 | 12 | 68 | 3.54 | ||||||
| Anterior cingulate | L | −5 | 19 | 28 | 26 | 3.52 | ||||||
| Middle temporal gyrus (22) | R | 59 | −33 | 4 | 80 | 3.41 | ||||||
| Parahippocampal gyrus | L | −23 | −35 | 4 | 6 | 3.39 | ||||||
| Posterior cingulate gyrus | L | −1 | −31 | 26 | 4 | 3.29 | ||||||
| S2 more than Awake | ||||||||||||
| IFG | L | −29 | 27 | −17 | 8 | 3.79 | ||||||
| Anatomical region (Brodmann area) . | Hemisphere . | x . | y . | z . | No. voxels . | Z-value . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Awake more than S2 | ||||||||||||
| STG (42) | R | 61 | −41 | 20 | 1404 | 4.85 | ||||||
| Cingulate gyrus (24, 32) | R | 9 | 9 | 46 | 3476 | 4.66 | ||||||
| L | −9 | 3 | 45 | 2404 | 4.2 | |||||||
| 0 | −29 | 31 | 116 | 3.92 | ||||||||
| MedFG | R | 9 | 9 | 47 | 1084 | 4.66 | ||||||
| L | −7 | 7 | 47 | 146 | 4.05 | |||||||
| Thalamus, anterior nucleus | R | 7 | −7 | 17 | 368 | 4.45 | ||||||
| R | 7 | −9 | 17 | 236 | 4.41 | |||||||
| L | −5 | −3 | 14 | 2 | 3.29 | |||||||
| Mammillary body/hypothalamus | L | −5 | −9 | −7 | 406 | 4.11 | ||||||
| Precentral gyrus | R | 37 | −7 | 41 | 230 | 4.36 | ||||||
| L | −47 | −7 | 49 | 288 | 3.54 | |||||||
| IPL (40) | R | 61 | −41 | 23 | 508 | 4.24 | ||||||
| SFG | R | 27 | 55 | −5 | 1024 | 4.2 | ||||||
| MFG | R | 27 | 55 | −6 | 608 | 4.2 | ||||||
| Insula | R | 39 | 13 | 3 | 792 | 4.05 | ||||||
| Post-central gyrus | R | 67 | −29 | 17 | 188 | 4.03 | ||||||
| Caudate | L | −23 | −37 | 9 | 87 | 3.92 | ||||||
| R | 5 | 1 | 14 | 78 | 3.77 | |||||||
| IFG (45) | R | 47 | 17 | 5 | 186 | 3.79 | ||||||
| Paracentral lobule | R | 7 | −19 | 45 | 196 | 3.63 | ||||||
| Transverse temporal gyrus | R | 51 | −27 | 12 | 68 | 3.54 | ||||||
| Anterior cingulate | L | −5 | 19 | 28 | 26 | 3.52 | ||||||
| Middle temporal gyrus (22) | R | 59 | −33 | 4 | 80 | 3.41 | ||||||
| Parahippocampal gyrus | L | −23 | −35 | 4 | 6 | 3.39 | ||||||
| Posterior cingulate gyrus | L | −1 | −31 | 26 | 4 | 3.29 | ||||||
| S2 more than Awake | ||||||||||||
| IFG | L | −29 | 27 | −17 | 8 | 3.79 | ||||||
Location of activated voxels in stereotactic space (MNI: x y z) for the comparison of Awake versus Sleep stage 2 (S2)
| Anatomical region (Brodmann area) . | Hemisphere . | x . | y . | z . | No. voxels . | Z-value . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Awake more than S2 | ||||||||||||
| STG (42) | R | 61 | −41 | 20 | 1404 | 4.85 | ||||||
| Cingulate gyrus (24, 32) | R | 9 | 9 | 46 | 3476 | 4.66 | ||||||
| L | −9 | 3 | 45 | 2404 | 4.2 | |||||||
| 0 | −29 | 31 | 116 | 3.92 | ||||||||
| MedFG | R | 9 | 9 | 47 | 1084 | 4.66 | ||||||
| L | −7 | 7 | 47 | 146 | 4.05 | |||||||
| Thalamus, anterior nucleus | R | 7 | −7 | 17 | 368 | 4.45 | ||||||
| R | 7 | −9 | 17 | 236 | 4.41 | |||||||
| L | −5 | −3 | 14 | 2 | 3.29 | |||||||
| Mammillary body/hypothalamus | L | −5 | −9 | −7 | 406 | 4.11 | ||||||
| Precentral gyrus | R | 37 | −7 | 41 | 230 | 4.36 | ||||||
| L | −47 | −7 | 49 | 288 | 3.54 | |||||||
| IPL (40) | R | 61 | −41 | 23 | 508 | 4.24 | ||||||
| SFG | R | 27 | 55 | −5 | 1024 | 4.2 | ||||||
| MFG | R | 27 | 55 | −6 | 608 | 4.2 | ||||||
| Insula | R | 39 | 13 | 3 | 792 | 4.05 | ||||||
| Post-central gyrus | R | 67 | −29 | 17 | 188 | 4.03 | ||||||
| Caudate | L | −23 | −37 | 9 | 87 | 3.92 | ||||||
| R | 5 | 1 | 14 | 78 | 3.77 | |||||||
| IFG (45) | R | 47 | 17 | 5 | 186 | 3.79 | ||||||
| Paracentral lobule | R | 7 | −19 | 45 | 196 | 3.63 | ||||||
| Transverse temporal gyrus | R | 51 | −27 | 12 | 68 | 3.54 | ||||||
| Anterior cingulate | L | −5 | 19 | 28 | 26 | 3.52 | ||||||
| Middle temporal gyrus (22) | R | 59 | −33 | 4 | 80 | 3.41 | ||||||
| Parahippocampal gyrus | L | −23 | −35 | 4 | 6 | 3.39 | ||||||
| Posterior cingulate gyrus | L | −1 | −31 | 26 | 4 | 3.29 | ||||||
| S2 more than Awake | ||||||||||||
| IFG | L | −29 | 27 | −17 | 8 | 3.79 | ||||||
| Anatomical region (Brodmann area) . | Hemisphere . | x . | y . | z . | No. voxels . | Z-value . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Awake more than S2 | ||||||||||||
| STG (42) | R | 61 | −41 | 20 | 1404 | 4.85 | ||||||
| Cingulate gyrus (24, 32) | R | 9 | 9 | 46 | 3476 | 4.66 | ||||||
| L | −9 | 3 | 45 | 2404 | 4.2 | |||||||
| 0 | −29 | 31 | 116 | 3.92 | ||||||||
| MedFG | R | 9 | 9 | 47 | 1084 | 4.66 | ||||||
| L | −7 | 7 | 47 | 146 | 4.05 | |||||||
| Thalamus, anterior nucleus | R | 7 | −7 | 17 | 368 | 4.45 | ||||||
| R | 7 | −9 | 17 | 236 | 4.41 | |||||||
| L | −5 | −3 | 14 | 2 | 3.29 | |||||||
| Mammillary body/hypothalamus | L | −5 | −9 | −7 | 406 | 4.11 | ||||||
| Precentral gyrus | R | 37 | −7 | 41 | 230 | 4.36 | ||||||
| L | −47 | −7 | 49 | 288 | 3.54 | |||||||
| IPL (40) | R | 61 | −41 | 23 | 508 | 4.24 | ||||||
| SFG | R | 27 | 55 | −5 | 1024 | 4.2 | ||||||
| MFG | R | 27 | 55 | −6 | 608 | 4.2 | ||||||
| Insula | R | 39 | 13 | 3 | 792 | 4.05 | ||||||
| Post-central gyrus | R | 67 | −29 | 17 | 188 | 4.03 | ||||||
| Caudate | L | −23 | −37 | 9 | 87 | 3.92 | ||||||
| R | 5 | 1 | 14 | 78 | 3.77 | |||||||
| IFG (45) | R | 47 | 17 | 5 | 186 | 3.79 | ||||||
| Paracentral lobule | R | 7 | −19 | 45 | 196 | 3.63 | ||||||
| Transverse temporal gyrus | R | 51 | −27 | 12 | 68 | 3.54 | ||||||
| Anterior cingulate | L | −5 | 19 | 28 | 26 | 3.52 | ||||||
| Middle temporal gyrus (22) | R | 59 | −33 | 4 | 80 | 3.41 | ||||||
| Parahippocampal gyrus | L | −23 | −35 | 4 | 6 | 3.39 | ||||||
| Posterior cingulate gyrus | L | −1 | −31 | 26 | 4 | 3.29 | ||||||
| S2 more than Awake | ||||||||||||
| IFG | L | −29 | 27 | −17 | 8 | 3.79 | ||||||
BOLD activity during wakefulness more than during SWS
During SWS deactivations were most prominent in frontal areas (MFG, SFG, MedFG, IFG and precentral gyrus), the cingulate cortex, in several regions of the association cortices (insular, temporal, parietal and occipital) and in subcortical structures such as the thalamus and hypothalamus. In more detail, during SWS there was bilaterally less activity in several regions of the frontal lobes (MedFG, MFG, SFG—BA22/adjacent to the insular cortex, IFG, precentral gyrus), the limbic lobe (ACC, cingulate gyrus, thalamus, caudate nucleus, parahippocampal gyrus, hippocampus, hypothalamus), the parietal lobes (supramarginal gyrus, IPL), the temporal lobes (MTG, STG, IFG, transverse temporal gyrus), the insula and the occipital cortex (precuneus, cuneus) (Table 4 and Fig. 1).
Location of activated voxels in stereotactic space (MNI: x y z) for the comparison of Awake versus Sleep stages 3/4 (SWS)
| Anatomical region (Brodmann area) . | Hemisphere . | x . | y . | z . | No. voxels . | Z-value . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Awake more than SWS | ||||||||||||
| MFG | R | 41 | 11 | 49 | 4194 | 6.37 | ||||||
| L | −39 | −1 | 49 | 4364 | 5.3 | |||||||
| L | −46 | 24 | 23 | 1169 | 5.15 | |||||||
| Cingulate gyrus (32) | L | −5 | −5 | 41 | 4214 | 5.75 | ||||||
| R | 9 | 11 | 46 | 7220 | 5.3 | |||||||
| 0 | −3 | 39 | 1469 | 4.57 | ||||||||
| 0 | −33 | 31 | 258 | 4.35 | ||||||||
| SFG (22) | R | 27 | 31 | 53 | 6094 | 5.42 | ||||||
| L | −21 | 53 | −3 | 1319 | 4.82 | |||||||
| 0 | 23 | 57 | 31 | 3.52 | ||||||||
| MedFG | R | 9 | 11 | 47 | 4336 | 5.3 | ||||||
| L | −20 | 53 | −3 | 1784 | 4.82 | |||||||
| 0 | 41 | 23 | 44 | 3.7 | ||||||||
| Precuneus | R | 21 | −75 | 35 | 1780 | 5.3 | ||||||
| IFG | L | −43 | 19 | −7 | 1842 | 5.17 | ||||||
| R | 41 | 15 | −13 | 432 | 3.92 | |||||||
| Supramarginal gyrus | L | −61 | −47 | 29 | 538 | 4.95 | ||||||
| R | 33 | −53 | 36 | 28 | 4.2 | |||||||
| Anterior cingulate (24) | L | −3 | 21 | 28 | 1218 | 4.85 | ||||||
| R | 17 | 37 | 20 | 2328 | 4.7 | |||||||
| 0 | 21 | 24 | 56 | 3.97 | ||||||||
| Precentral gyrus | L | −39 | −1 | 46 | 2296 | 4.7 | ||||||
| R | 37 | −5 | 41 | 682 | 4.37 | |||||||
| IPL | L | −51 | −35 | 29 | 486 | 4.6 | ||||||
| R | 35 | −59 | 43 | 2192 | 4.5 | |||||||
| Insula | L | −27 | −21 | 22 | 1942 | 4.6 | ||||||
| R | 57 | −35 | 19 | 754 | 4.47 | |||||||
| Middle temporal gyrus | L | −45 | −59 | 5 | 828 | 4.6 | ||||||
| R | 65 | −33 | 3 | 1214 | 4.4 | |||||||
| Thalamus, anterior nucleus | R | 7 | −7 | 18 | 482 | 4.3 | ||||||
| R | 7 | −9 | 18 | 52 | 3.87 | |||||||
| STG | L | −45 | −7 | −1 | 863 | 4.55 | ||||||
| R | 47 | 11 | −13 | 1816 | 4.32 | |||||||
| Caudate | L | −14 | −3 | 22 | 464 | 4.42 | ||||||
| Cuneus | R | 19 | −77 | 32 | 316 | 4.27 | ||||||
| Paracentral lobule | R | 9 | −11 | 45 | 136 | 4.42 | ||||||
| Superior parietal lobule | R | 31 | −61 | 43 | 120 | 4.1 | ||||||
| Transverse temporal gyrus | L | −43 | −21 | 12 | 110 | 4.07 | ||||||
| Posterior cingulate gyrus | R | 5 | −47 | 19 | 160 | 4.02 | ||||||
| L | −1 | −31 | 26 | 192 | 3.55 | |||||||
| Post-central gyrus | R | 53 | −29 | 17 | 44 | 3.75 | ||||||
| L | −47 | −17 | 13 | 84 | 3.65 | |||||||
| Angular gyrus | R | 33 | −55 | 36 | 292 | 3.77 | ||||||
| Subcallosal gyrus | R | 25 | 8 | −14 | 66 | 3.7 | ||||||
| Claustrum | L | −26 | −23 | 16 | 22 | 3.7 | ||||||
| Parahippocampal gyrus (30/34) | R | 27 | 5 | −18 | 236 | 3.67 | ||||||
| L | −24 | −35 | 4 | 22 | 3.42 | |||||||
| Mammillary body/hypothalamus | 0 | −9 | −5 | 24 | 3.67 | |||||||
| Uncus | R | 27 | 5 | −22 | 4 | 3.17 | ||||||
| SWS more than Awake | ||||||||||||
| STG (38, 22) | L | −51 | 21 | −23 | 97 | 4.71 | ||||||
| R | 63 | −3 | 3 | 352 | 4.32 | |||||||
| Parahippocampal gyrus | R | 19 | −15 | −11 | 289 | 4.56 | ||||||
| R | 17 | −15 | −14 | 88 | 4.45 | |||||||
| Declive | L | −41 | −71 | −27 | 236 | 4.37 | ||||||
| IFG | L | −34 | 34 | −15 | 116 | 4.26 | ||||||
| MFG | L | −35 | 35 | −15 | 92 | 4.26 | ||||||
| Inferior temporal gyrus | L | −63 | −13 | −19 | 104 | 4.17 | ||||||
| Culmen | L | −17 | −49 | −29 | 22 | 4.17 | ||||||
| Transverse temporal gyrus | R | 63 | −7 | 9 | 60 | 4.06 | ||||||
| Middle temporal gyrus | L | −51 | −23 | −9 | 50 | 4.06 | ||||||
| Fusiform gyrus | L | −41 | −73 | −21 | 32 | 3.99 | ||||||
| Precentral gyrus | R | 61 | −7 | 11 | 48 | 3.91 | ||||||
| Post-central gyrus | R | 63 | −7 | 15 | 8 | 3.86 | ||||||
| Middle occipital gyrus | L | −45 | −75 | −17 | 4 | 3.6 | ||||||
| Anatomical region (Brodmann area) . | Hemisphere . | x . | y . | z . | No. voxels . | Z-value . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Awake more than SWS | ||||||||||||
| MFG | R | 41 | 11 | 49 | 4194 | 6.37 | ||||||
| L | −39 | −1 | 49 | 4364 | 5.3 | |||||||
| L | −46 | 24 | 23 | 1169 | 5.15 | |||||||
| Cingulate gyrus (32) | L | −5 | −5 | 41 | 4214 | 5.75 | ||||||
| R | 9 | 11 | 46 | 7220 | 5.3 | |||||||
| 0 | −3 | 39 | 1469 | 4.57 | ||||||||
| 0 | −33 | 31 | 258 | 4.35 | ||||||||
| SFG (22) | R | 27 | 31 | 53 | 6094 | 5.42 | ||||||
| L | −21 | 53 | −3 | 1319 | 4.82 | |||||||
| 0 | 23 | 57 | 31 | 3.52 | ||||||||
| MedFG | R | 9 | 11 | 47 | 4336 | 5.3 | ||||||
| L | −20 | 53 | −3 | 1784 | 4.82 | |||||||
| 0 | 41 | 23 | 44 | 3.7 | ||||||||
| Precuneus | R | 21 | −75 | 35 | 1780 | 5.3 | ||||||
| IFG | L | −43 | 19 | −7 | 1842 | 5.17 | ||||||
| R | 41 | 15 | −13 | 432 | 3.92 | |||||||
| Supramarginal gyrus | L | −61 | −47 | 29 | 538 | 4.95 | ||||||
| R | 33 | −53 | 36 | 28 | 4.2 | |||||||
| Anterior cingulate (24) | L | −3 | 21 | 28 | 1218 | 4.85 | ||||||
| R | 17 | 37 | 20 | 2328 | 4.7 | |||||||
| 0 | 21 | 24 | 56 | 3.97 | ||||||||
| Precentral gyrus | L | −39 | −1 | 46 | 2296 | 4.7 | ||||||
| R | 37 | −5 | 41 | 682 | 4.37 | |||||||
| IPL | L | −51 | −35 | 29 | 486 | 4.6 | ||||||
| R | 35 | −59 | 43 | 2192 | 4.5 | |||||||
| Insula | L | −27 | −21 | 22 | 1942 | 4.6 | ||||||
| R | 57 | −35 | 19 | 754 | 4.47 | |||||||
| Middle temporal gyrus | L | −45 | −59 | 5 | 828 | 4.6 | ||||||
| R | 65 | −33 | 3 | 1214 | 4.4 | |||||||
| Thalamus, anterior nucleus | R | 7 | −7 | 18 | 482 | 4.3 | ||||||
| R | 7 | −9 | 18 | 52 | 3.87 | |||||||
| STG | L | −45 | −7 | −1 | 863 | 4.55 | ||||||
| R | 47 | 11 | −13 | 1816 | 4.32 | |||||||
| Caudate | L | −14 | −3 | 22 | 464 | 4.42 | ||||||
| Cuneus | R | 19 | −77 | 32 | 316 | 4.27 | ||||||
| Paracentral lobule | R | 9 | −11 | 45 | 136 | 4.42 | ||||||
| Superior parietal lobule | R | 31 | −61 | 43 | 120 | 4.1 | ||||||
| Transverse temporal gyrus | L | −43 | −21 | 12 | 110 | 4.07 | ||||||
| Posterior cingulate gyrus | R | 5 | −47 | 19 | 160 | 4.02 | ||||||
| L | −1 | −31 | 26 | 192 | 3.55 | |||||||
| Post-central gyrus | R | 53 | −29 | 17 | 44 | 3.75 | ||||||
| L | −47 | −17 | 13 | 84 | 3.65 | |||||||
| Angular gyrus | R | 33 | −55 | 36 | 292 | 3.77 | ||||||
| Subcallosal gyrus | R | 25 | 8 | −14 | 66 | 3.7 | ||||||
| Claustrum | L | −26 | −23 | 16 | 22 | 3.7 | ||||||
| Parahippocampal gyrus (30/34) | R | 27 | 5 | −18 | 236 | 3.67 | ||||||
| L | −24 | −35 | 4 | 22 | 3.42 | |||||||
| Mammillary body/hypothalamus | 0 | −9 | −5 | 24 | 3.67 | |||||||
| Uncus | R | 27 | 5 | −22 | 4 | 3.17 | ||||||
| SWS more than Awake | ||||||||||||
| STG (38, 22) | L | −51 | 21 | −23 | 97 | 4.71 | ||||||
| R | 63 | −3 | 3 | 352 | 4.32 | |||||||
| Parahippocampal gyrus | R | 19 | −15 | −11 | 289 | 4.56 | ||||||
| R | 17 | −15 | −14 | 88 | 4.45 | |||||||
| Declive | L | −41 | −71 | −27 | 236 | 4.37 | ||||||
| IFG | L | −34 | 34 | −15 | 116 | 4.26 | ||||||
| MFG | L | −35 | 35 | −15 | 92 | 4.26 | ||||||
| Inferior temporal gyrus | L | −63 | −13 | −19 | 104 | 4.17 | ||||||
| Culmen | L | −17 | −49 | −29 | 22 | 4.17 | ||||||
| Transverse temporal gyrus | R | 63 | −7 | 9 | 60 | 4.06 | ||||||
| Middle temporal gyrus | L | −51 | −23 | −9 | 50 | 4.06 | ||||||
| Fusiform gyrus | L | −41 | −73 | −21 | 32 | 3.99 | ||||||
| Precentral gyrus | R | 61 | −7 | 11 | 48 | 3.91 | ||||||
| Post-central gyrus | R | 63 | −7 | 15 | 8 | 3.86 | ||||||
| Middle occipital gyrus | L | −45 | −75 | −17 | 4 | 3.6 | ||||||
Location of activated voxels in stereotactic space (MNI: x y z) for the comparison of Awake versus Sleep stages 3/4 (SWS)
| Anatomical region (Brodmann area) . | Hemisphere . | x . | y . | z . | No. voxels . | Z-value . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Awake more than SWS | ||||||||||||
| MFG | R | 41 | 11 | 49 | 4194 | 6.37 | ||||||
| L | −39 | −1 | 49 | 4364 | 5.3 | |||||||
| L | −46 | 24 | 23 | 1169 | 5.15 | |||||||
| Cingulate gyrus (32) | L | −5 | −5 | 41 | 4214 | 5.75 | ||||||
| R | 9 | 11 | 46 | 7220 | 5.3 | |||||||
| 0 | −3 | 39 | 1469 | 4.57 | ||||||||
| 0 | −33 | 31 | 258 | 4.35 | ||||||||
| SFG (22) | R | 27 | 31 | 53 | 6094 | 5.42 | ||||||
| L | −21 | 53 | −3 | 1319 | 4.82 | |||||||
| 0 | 23 | 57 | 31 | 3.52 | ||||||||
| MedFG | R | 9 | 11 | 47 | 4336 | 5.3 | ||||||
| L | −20 | 53 | −3 | 1784 | 4.82 | |||||||
| 0 | 41 | 23 | 44 | 3.7 | ||||||||
| Precuneus | R | 21 | −75 | 35 | 1780 | 5.3 | ||||||
| IFG | L | −43 | 19 | −7 | 1842 | 5.17 | ||||||
| R | 41 | 15 | −13 | 432 | 3.92 | |||||||
| Supramarginal gyrus | L | −61 | −47 | 29 | 538 | 4.95 | ||||||
| R | 33 | −53 | 36 | 28 | 4.2 | |||||||
| Anterior cingulate (24) | L | −3 | 21 | 28 | 1218 | 4.85 | ||||||
| R | 17 | 37 | 20 | 2328 | 4.7 | |||||||
| 0 | 21 | 24 | 56 | 3.97 | ||||||||
| Precentral gyrus | L | −39 | −1 | 46 | 2296 | 4.7 | ||||||
| R | 37 | −5 | 41 | 682 | 4.37 | |||||||
| IPL | L | −51 | −35 | 29 | 486 | 4.6 | ||||||
| R | 35 | −59 | 43 | 2192 | 4.5 | |||||||
| Insula | L | −27 | −21 | 22 | 1942 | 4.6 | ||||||
| R | 57 | −35 | 19 | 754 | 4.47 | |||||||
| Middle temporal gyrus | L | −45 | −59 | 5 | 828 | 4.6 | ||||||
| R | 65 | −33 | 3 | 1214 | 4.4 | |||||||
| Thalamus, anterior nucleus | R | 7 | −7 | 18 | 482 | 4.3 | ||||||
| R | 7 | −9 | 18 | 52 | 3.87 | |||||||
| STG | L | −45 | −7 | −1 | 863 | 4.55 | ||||||
| R | 47 | 11 | −13 | 1816 | 4.32 | |||||||
| Caudate | L | −14 | −3 | 22 | 464 | 4.42 | ||||||
| Cuneus | R | 19 | −77 | 32 | 316 | 4.27 | ||||||
| Paracentral lobule | R | 9 | −11 | 45 | 136 | 4.42 | ||||||
| Superior parietal lobule | R | 31 | −61 | 43 | 120 | 4.1 | ||||||
| Transverse temporal gyrus | L | −43 | −21 | 12 | 110 | 4.07 | ||||||
| Posterior cingulate gyrus | R | 5 | −47 | 19 | 160 | 4.02 | ||||||
| L | −1 | −31 | 26 | 192 | 3.55 | |||||||
| Post-central gyrus | R | 53 | −29 | 17 | 44 | 3.75 | ||||||
| L | −47 | −17 | 13 | 84 | 3.65 | |||||||
| Angular gyrus | R | 33 | −55 | 36 | 292 | 3.77 | ||||||
| Subcallosal gyrus | R | 25 | 8 | −14 | 66 | 3.7 | ||||||
| Claustrum | L | −26 | −23 | 16 | 22 | 3.7 | ||||||
| Parahippocampal gyrus (30/34) | R | 27 | 5 | −18 | 236 | 3.67 | ||||||
| L | −24 | −35 | 4 | 22 | 3.42 | |||||||
| Mammillary body/hypothalamus | 0 | −9 | −5 | 24 | 3.67 | |||||||
| Uncus | R | 27 | 5 | −22 | 4 | 3.17 | ||||||
| SWS more than Awake | ||||||||||||
| STG (38, 22) | L | −51 | 21 | −23 | 97 | 4.71 | ||||||
| R | 63 | −3 | 3 | 352 | 4.32 | |||||||
| Parahippocampal gyrus | R | 19 | −15 | −11 | 289 | 4.56 | ||||||
| R | 17 | −15 | −14 | 88 | 4.45 | |||||||
| Declive | L | −41 | −71 | −27 | 236 | 4.37 | ||||||
| IFG | L | −34 | 34 | −15 | 116 | 4.26 | ||||||
| MFG | L | −35 | 35 | −15 | 92 | 4.26 | ||||||
| Inferior temporal gyrus | L | −63 | −13 | −19 | 104 | 4.17 | ||||||
| Culmen | L | −17 | −49 | −29 | 22 | 4.17 | ||||||
| Transverse temporal gyrus | R | 63 | −7 | 9 | 60 | 4.06 | ||||||
| Middle temporal gyrus | L | −51 | −23 | −9 | 50 | 4.06 | ||||||
| Fusiform gyrus | L | −41 | −73 | −21 | 32 | 3.99 | ||||||
| Precentral gyrus | R | 61 | −7 | 11 | 48 | 3.91 | ||||||
| Post-central gyrus | R | 63 | −7 | 15 | 8 | 3.86 | ||||||
| Middle occipital gyrus | L | −45 | −75 | −17 | 4 | 3.6 | ||||||
| Anatomical region (Brodmann area) . | Hemisphere . | x . | y . | z . | No. voxels . | Z-value . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Awake more than SWS | ||||||||||||
| MFG | R | 41 | 11 | 49 | 4194 | 6.37 | ||||||
| L | −39 | −1 | 49 | 4364 | 5.3 | |||||||
| L | −46 | 24 | 23 | 1169 | 5.15 | |||||||
| Cingulate gyrus (32) | L | −5 | −5 | 41 | 4214 | 5.75 | ||||||
| R | 9 | 11 | 46 | 7220 | 5.3 | |||||||
| 0 | −3 | 39 | 1469 | 4.57 | ||||||||
| 0 | −33 | 31 | 258 | 4.35 | ||||||||
| SFG (22) | R | 27 | 31 | 53 | 6094 | 5.42 | ||||||
| L | −21 | 53 | −3 | 1319 | 4.82 | |||||||
| 0 | 23 | 57 | 31 | 3.52 | ||||||||
| MedFG | R | 9 | 11 | 47 | 4336 | 5.3 | ||||||
| L | −20 | 53 | −3 | 1784 | 4.82 | |||||||
| 0 | 41 | 23 | 44 | 3.7 | ||||||||
| Precuneus | R | 21 | −75 | 35 | 1780 | 5.3 | ||||||
| IFG | L | −43 | 19 | −7 | 1842 | 5.17 | ||||||
| R | 41 | 15 | −13 | 432 | 3.92 | |||||||
| Supramarginal gyrus | L | −61 | −47 | 29 | 538 | 4.95 | ||||||
| R | 33 | −53 | 36 | 28 | 4.2 | |||||||
| Anterior cingulate (24) | L | −3 | 21 | 28 | 1218 | 4.85 | ||||||
| R | 17 | 37 | 20 | 2328 | 4.7 | |||||||
| 0 | 21 | 24 | 56 | 3.97 | ||||||||
| Precentral gyrus | L | −39 | −1 | 46 | 2296 | 4.7 | ||||||
| R | 37 | −5 | 41 | 682 | 4.37 | |||||||
| IPL | L | −51 | −35 | 29 | 486 | 4.6 | ||||||
| R | 35 | −59 | 43 | 2192 | 4.5 | |||||||
| Insula | L | −27 | −21 | 22 | 1942 | 4.6 | ||||||
| R | 57 | −35 | 19 | 754 | 4.47 | |||||||
| Middle temporal gyrus | L | −45 | −59 | 5 | 828 | 4.6 | ||||||
| R | 65 | −33 | 3 | 1214 | 4.4 | |||||||
| Thalamus, anterior nucleus | R | 7 | −7 | 18 | 482 | 4.3 | ||||||
| R | 7 | −9 | 18 | 52 | 3.87 | |||||||
| STG | L | −45 | −7 | −1 | 863 | 4.55 | ||||||
| R | 47 | 11 | −13 | 1816 | 4.32 | |||||||
| Caudate | L | −14 | −3 | 22 | 464 | 4.42 | ||||||
| Cuneus | R | 19 | −77 | 32 | 316 | 4.27 | ||||||
| Paracentral lobule | R | 9 | −11 | 45 | 136 | 4.42 | ||||||
| Superior parietal lobule | R | 31 | −61 | 43 | 120 | 4.1 | ||||||
| Transverse temporal gyrus | L | −43 | −21 | 12 | 110 | 4.07 | ||||||
| Posterior cingulate gyrus | R | 5 | −47 | 19 | 160 | 4.02 | ||||||
| L | −1 | −31 | 26 | 192 | 3.55 | |||||||
| Post-central gyrus | R | 53 | −29 | 17 | 44 | 3.75 | ||||||
| L | −47 | −17 | 13 | 84 | 3.65 | |||||||
| Angular gyrus | R | 33 | −55 | 36 | 292 | 3.77 | ||||||
| Subcallosal gyrus | R | 25 | 8 | −14 | 66 | 3.7 | ||||||
| Claustrum | L | −26 | −23 | 16 | 22 | 3.7 | ||||||
| Parahippocampal gyrus (30/34) | R | 27 | 5 | −18 | 236 | 3.67 | ||||||
| L | −24 | −35 | 4 | 22 | 3.42 | |||||||
| Mammillary body/hypothalamus | 0 | −9 | −5 | 24 | 3.67 | |||||||
| Uncus | R | 27 | 5 | −22 | 4 | 3.17 | ||||||
| SWS more than Awake | ||||||||||||
| STG (38, 22) | L | −51 | 21 | −23 | 97 | 4.71 | ||||||
| R | 63 | −3 | 3 | 352 | 4.32 | |||||||
| Parahippocampal gyrus | R | 19 | −15 | −11 | 289 | 4.56 | ||||||
| R | 17 | −15 | −14 | 88 | 4.45 | |||||||
| Declive | L | −41 | −71 | −27 | 236 | 4.37 | ||||||
| IFG | L | −34 | 34 | −15 | 116 | 4.26 | ||||||
| MFG | L | −35 | 35 | −15 | 92 | 4.26 | ||||||
| Inferior temporal gyrus | L | −63 | −13 | −19 | 104 | 4.17 | ||||||
| Culmen | L | −17 | −49 | −29 | 22 | 4.17 | ||||||
| Transverse temporal gyrus | R | 63 | −7 | 9 | 60 | 4.06 | ||||||
| Middle temporal gyrus | L | −51 | −23 | −9 | 50 | 4.06 | ||||||
| Fusiform gyrus | L | −41 | −73 | −21 | 32 | 3.99 | ||||||
| Precentral gyrus | R | 61 | −7 | 11 | 48 | 3.91 | ||||||
| Post-central gyrus | R | 63 | −7 | 15 | 8 | 3.86 | ||||||
| Middle occipital gyrus | L | −45 | −75 | −17 | 4 | 3.6 | ||||||
BOLD activity during wakefulness less than during stage 1
Compared with wakefulness we found (small) occipital (precuneus) and temporal regions to be more active but the number of activated voxels was considerably smaller than that of the opposite contrast (Table 2 and Fig. 1).
BOLD activity during wakefulness less than during stage 2
During S2 there was more activity only in a small region of left IFG (Table 3 and Fig. 1).
BOLD activity during wakefulness less than during SWS
SWS showed more activity in temporal, parahippocampal and cerebellar regions. More precisely, activations were related to the STG, the parahippocampal gyrus, the cerebellum (declive, culmen), and the small regions of the IFG, MFG, inferior temporal gyrus and fusiform gyrus (Table 4 and Fig. 1). Remarkably, there were considerably less voxels more active during sleep, e.g. for the comparison ‘Awake versus SWS’ there is a ratio of 97 to 3% of the degree of estimated voxel activity.
Comparing BOLD-related brain activity between NREM sleep stages
Stage 1 versus stage 2
S1 showed more BOLD-related activity than S2 in the middle and MedFG, supramarginal gyrus, superior temporal gyri, cingulate cortex (mainly median), supplementary motor area and paracentral lobule. In contrast, S2 showed more activity in the cerebellum, the parahippocampal gyrus and the hippocampus.
Stage 1 versus SWS
Compared with SWS S1 showed more activation in the anterior cingulate gyrus. Similar to S2, SWS showed more activation compared with S1 in the hippocampus, the parahippocampal gyrus and the cerebellum.
Stage 2 versus SWS
S2 showed more activity in the anterior cingulate gyrus when compared with SWS. SWS compared with S2 showed more activity in the middle occipital gyrus, cerebellum, parahippocampal gyrus, hippocampus, pre- and post-central gyrus, inferior temporal gyrus, angular gyrus, precuneus and right insula.
Comparing functional connectivity of hypothalamic activity
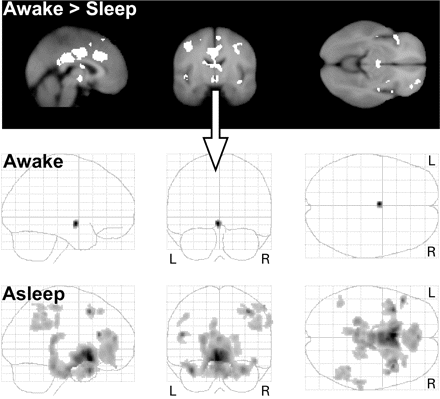
The hypothalamic region was less active throughout all NREM sleep stages (including S1) as compared with wakefulness. Calculating the functional connectivity of this region to any other brain area revealed several regions for the first sleep cycle periods: cingulate gyrus (anterior, median, posterior), caudate nucleus, frontal gyri (middle, inferior, superior, precentral), hippocampus, parahippocampal gyrus, thalamus (pulvinar), IPL, angular gyrus, temporal gyri (middle, inferior) and brainstem (pons, midbrain). In contrast, hypothalamic activity of only the awake period (close to sleep onset) revealed no connected regions (Table 5 and Fig. 2).
First row: Contrasting the BOLD response of resting awake with NREM sleep (S1 to SWS) with less smoothed data (4 mm FWHM) to enhance sensitivity for subcortical regions. We extracted the averaged hypothalamic time series (arrow) and modelled them as regressors of interest in a second SPM analysis to determine functional connectivity between the awake and sleeping conditions. Second and third rows: Functional connectivity map with hypothalamic regions (volume 5 × 10 × 3 mm at MNI coordinates −2, −10, −10). The grey coloured regions (Table 5) within the glass brains denote functional connectivity of hypothalamic activity during the resting awake state (shortly before sleep onset) and during NREM sleep when hypothalamic activity was decreased compared with the awake state.
Location of activated voxels in stereotactic space (MNI: x y z) for hypothalamic connectivity
| Anatomical region . | Hemisphere . | x . | y . | z . | Z-value . |
|---|---|---|---|---|---|
| Anterior cingulate gyrus | L | −4 | 14 | −12 | 5.07 |
| Caudate nucleus, head | R | 2 | 8 | −6 | 4.86 |
| Hypothalamus | R | 4 | 0 | −12 | 4.67 |
| MFG | R | 38 | 14 | 48 | 4.51 |
| L | −40 | 30 | 34 | 3.52 | |
| IFG, triangular part | L | −48 | 26 | 18 | 4.47 |
| R | 46 | 40 | 4 | 3.82 | |
| Hippocampus | L | −22 | −34 | 10 | 4.05 |
| Median cingulate gyrus | R | 12 | −48 | 36 | 3.89 |
| IPL | R | 58 | −32 | 56 | 3.84 |
| L | −44 | −54 | 42 | 3.5 | |
| Parahippocampal gyrus | L | −18 | −34 | −6 | 3.83 |
| R | 22 | −34 | −4 | 3.43 | |
| Middle temporal gyrus | L | −52 | −56 | 16 | 3.78 |
| Posterior cingulate gyrus | L | −6 | −42 | 32 | 3.73 |
| R | 10 | −48 | 14 | 3.46 | |
| Angular gyrus | R | 46 | −50 | 28 | 3.63 |
| L | −52 | −54 | 34 | 3.12 | |
| Inferior temporal gyrus | R | 36 | 2 | −42 | 3.6 |
| Precentral gyrus | R | 54 | −2 | 48 | 3.59 |
| L | −58 | −8 | 34 | 3.03 | |
| Thalamus, pulvinar | L | −16 | −24 | 18 | 3.54 |
| R | 20 | −30 | 14 | 3.53 | |
| Supramarginal gyrus | R | 66 | −28 | 36 | 3.53 |
| Post-central gyrus | L | −48 | −14 | 32 | 3.42 |
| SFG, dorsolateral | R | 26 | 30 | 50 | 3.3 |
| Brainstem, pons | R | 6 | −24 | −34 | 3.02 |
| Brainstem, midbrain | L | −2 | −15 | −28 | 2.91 |
| Anatomical region . | Hemisphere . | x . | y . | z . | Z-value . |
|---|---|---|---|---|---|
| Anterior cingulate gyrus | L | −4 | 14 | −12 | 5.07 |
| Caudate nucleus, head | R | 2 | 8 | −6 | 4.86 |
| Hypothalamus | R | 4 | 0 | −12 | 4.67 |
| MFG | R | 38 | 14 | 48 | 4.51 |
| L | −40 | 30 | 34 | 3.52 | |
| IFG, triangular part | L | −48 | 26 | 18 | 4.47 |
| R | 46 | 40 | 4 | 3.82 | |
| Hippocampus | L | −22 | −34 | 10 | 4.05 |
| Median cingulate gyrus | R | 12 | −48 | 36 | 3.89 |
| IPL | R | 58 | −32 | 56 | 3.84 |
| L | −44 | −54 | 42 | 3.5 | |
| Parahippocampal gyrus | L | −18 | −34 | −6 | 3.83 |
| R | 22 | −34 | −4 | 3.43 | |
| Middle temporal gyrus | L | −52 | −56 | 16 | 3.78 |
| Posterior cingulate gyrus | L | −6 | −42 | 32 | 3.73 |
| R | 10 | −48 | 14 | 3.46 | |
| Angular gyrus | R | 46 | −50 | 28 | 3.63 |
| L | −52 | −54 | 34 | 3.12 | |
| Inferior temporal gyrus | R | 36 | 2 | −42 | 3.6 |
| Precentral gyrus | R | 54 | −2 | 48 | 3.59 |
| L | −58 | −8 | 34 | 3.03 | |
| Thalamus, pulvinar | L | −16 | −24 | 18 | 3.54 |
| R | 20 | −30 | 14 | 3.53 | |
| Supramarginal gyrus | R | 66 | −28 | 36 | 3.53 |
| Post-central gyrus | L | −48 | −14 | 32 | 3.42 |
| SFG, dorsolateral | R | 26 | 30 | 50 | 3.3 |
| Brainstem, pons | R | 6 | −24 | −34 | 3.02 |
| Brainstem, midbrain | L | −2 | −15 | −28 | 2.91 |
Location of activated voxels in stereotactic space (MNI: x y z) for hypothalamic connectivity
| Anatomical region . | Hemisphere . | x . | y . | z . | Z-value . |
|---|---|---|---|---|---|
| Anterior cingulate gyrus | L | −4 | 14 | −12 | 5.07 |
| Caudate nucleus, head | R | 2 | 8 | −6 | 4.86 |
| Hypothalamus | R | 4 | 0 | −12 | 4.67 |
| MFG | R | 38 | 14 | 48 | 4.51 |
| L | −40 | 30 | 34 | 3.52 | |
| IFG, triangular part | L | −48 | 26 | 18 | 4.47 |
| R | 46 | 40 | 4 | 3.82 | |
| Hippocampus | L | −22 | −34 | 10 | 4.05 |
| Median cingulate gyrus | R | 12 | −48 | 36 | 3.89 |
| IPL | R | 58 | −32 | 56 | 3.84 |
| L | −44 | −54 | 42 | 3.5 | |
| Parahippocampal gyrus | L | −18 | −34 | −6 | 3.83 |
| R | 22 | −34 | −4 | 3.43 | |
| Middle temporal gyrus | L | −52 | −56 | 16 | 3.78 |
| Posterior cingulate gyrus | L | −6 | −42 | 32 | 3.73 |
| R | 10 | −48 | 14 | 3.46 | |
| Angular gyrus | R | 46 | −50 | 28 | 3.63 |
| L | −52 | −54 | 34 | 3.12 | |
| Inferior temporal gyrus | R | 36 | 2 | −42 | 3.6 |
| Precentral gyrus | R | 54 | −2 | 48 | 3.59 |
| L | −58 | −8 | 34 | 3.03 | |
| Thalamus, pulvinar | L | −16 | −24 | 18 | 3.54 |
| R | 20 | −30 | 14 | 3.53 | |
| Supramarginal gyrus | R | 66 | −28 | 36 | 3.53 |
| Post-central gyrus | L | −48 | −14 | 32 | 3.42 |
| SFG, dorsolateral | R | 26 | 30 | 50 | 3.3 |
| Brainstem, pons | R | 6 | −24 | −34 | 3.02 |
| Brainstem, midbrain | L | −2 | −15 | −28 | 2.91 |
| Anatomical region . | Hemisphere . | x . | y . | z . | Z-value . |
|---|---|---|---|---|---|
| Anterior cingulate gyrus | L | −4 | 14 | −12 | 5.07 |
| Caudate nucleus, head | R | 2 | 8 | −6 | 4.86 |
| Hypothalamus | R | 4 | 0 | −12 | 4.67 |
| MFG | R | 38 | 14 | 48 | 4.51 |
| L | −40 | 30 | 34 | 3.52 | |
| IFG, triangular part | L | −48 | 26 | 18 | 4.47 |
| R | 46 | 40 | 4 | 3.82 | |
| Hippocampus | L | −22 | −34 | 10 | 4.05 |
| Median cingulate gyrus | R | 12 | −48 | 36 | 3.89 |
| IPL | R | 58 | −32 | 56 | 3.84 |
| L | −44 | −54 | 42 | 3.5 | |
| Parahippocampal gyrus | L | −18 | −34 | −6 | 3.83 |
| R | 22 | −34 | −4 | 3.43 | |
| Middle temporal gyrus | L | −52 | −56 | 16 | 3.78 |
| Posterior cingulate gyrus | L | −6 | −42 | 32 | 3.73 |
| R | 10 | −48 | 14 | 3.46 | |
| Angular gyrus | R | 46 | −50 | 28 | 3.63 |
| L | −52 | −54 | 34 | 3.12 | |
| Inferior temporal gyrus | R | 36 | 2 | −42 | 3.6 |
| Precentral gyrus | R | 54 | −2 | 48 | 3.59 |
| L | −58 | −8 | 34 | 3.03 | |
| Thalamus, pulvinar | L | −16 | −24 | 18 | 3.54 |
| R | 20 | −30 | 14 | 3.53 | |
| Supramarginal gyrus | R | 66 | −28 | 36 | 3.53 |
| Post-central gyrus | L | −48 | −14 | 32 | 3.42 |
| SFG, dorsolateral | R | 26 | 30 | 50 | 3.3 |
| Brainstem, pons | R | 6 | −24 | −34 | 3.02 |
| Brainstem, midbrain | L | −2 | −15 | −28 | 2.91 |
Discussion
We simultaneously measured spontaneous EEG and fMRI during the night's first sleep cycles and report here, for the first time, NREM sleep stage and regional specific alterations of the BOLD response. When comparing light sleep (S1 and S2) with wakefulness brain activity was considerably decreased in all lobes of the cerebrum, the cingulate cortex, the insula and the thalamus, whereas it was further reduced during SWS in the ACC, left insula and hippocampal regions.
Our results are in overall agreement with earlier data from other laboratories employing different methodologies: these studies used PET to relate measurements of either cerebral blood flow or cerebral glucose utilization with light sleep (Kajimura et al., 1999), S1 (Kjaer et al., 2002), S2 (Andersson et al., 1998; Maquet et al., 1992) or with a ‘mix’ of NREM sleep (Nofzinger et al., 2002), or to compare SWS with an alert awake state (Buchsbaum et al., 1989; Maquet et al., 1990, 1997; Braun et al., 1997; Hofle et al., 1997; Kajimura et al., 1999). All studies found less blood flow or metabolism in the thalamus and in the cortex (with varying locations) during sleep, but the different intrinsic temporal resolution and baseline references hamper an easy comparison.
Topography of reduced brain activity during NREM sleep
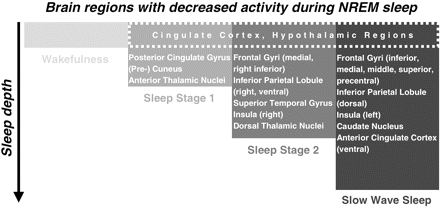
To sum up our sleep stage specific findings we propose a topography of reduced activity from S1 to SWS (Fig. 3) as follows. In comparison with S2 and SWS there is less activation related to S1 within the PCG (BA 23), the cuneus and precuneus, and the thalamic nuclei. Brain regions deactivated during S2 are the MedFG (BA 6), the right ventral part of the IPL (BA 40), the STG, the right insula (BA 13) and the right IFG (BA 45). In addition, there are several regions less activated during SWS: the frontal gyri (BA 6, 8, 9 and 45), the precentral gyrus, bilaterally the dorsal IPL (BA 40), the cingulate gyri (BA 23, 24, 32), the left insula (BA 13), the caudate body, the hippocampus and the parahippocampal gyri (BA 34). Following these results we suggest a network of cortical brain regions relevant for falling asleep. When falling asleep (S1) occipital, cingulate, posterior cingulate, thalamic and hypothalamic regions reduce their activity. Subsequently, when sleep gets synchronized (S2) most of these regions remain less activated with the exception of the thalamic nuclei and occipital regions. But in addition the activity of the right insula (anterior and posterior) as well as frontal, temporal and parietal regions is reduced. Finally, when reaching SWS the activity of the ACC, left insula and hippocampal regions is decreased.
A schematic illustration of brain regions with decreased activity during different stages of NREM sleep. The model is a summary derived from our results and we expect that the precise anatomical localizations may vary depending on pre-sleep activity. Regions of the cingulate cortex and hypothalamic regions alter their activity in all NREM sleep stages (box with dashed line). As sleep deepens different brain regions get involved indicating that NREM sleep is associated with dynamic regional brain processes with sleep stage specific activation and deactivation patterns as indicated.
It has been proposed that at any time some neuronal groups are in a so-called disjunctive state, i.e. in a sleeping-like state due to a temporary disjunction at the local (neuronal) level between input and output of a neuronal group (Krueger et al., 1995). If a sufficient number of neuronal groups are in the disjunctive state, the discontinuation of the perception of wakefulness occurs. As this is usually linked to S2 we suggest that the network for a disjunctive state might comprise thalamic and hypothalamic regions, the cingulate cortex, the right insula, the IPL, STG, and IFG as well as MFG. Therefore, it is unlikely that the establishment of synchronized sleep can be explained by a single ‘superstructure’. Additionally, our model is contradictory to that of Kajimura et al., 1999. They suggested three groups of brain structures each representing one type of deactivation during the progression of NREM sleep. Group one comprises brain regions deactivated during sleep irrespective of sleep stages (cerebellum, putamen, ACC, IFG, MFG and IPL). Group two and three refer to brain structures specific for light sleep (pons, thalamus) and deep sleep (midbrain, caudate nucleus, vermis). Our observations differ concerning the role of the cerebellum, the insula, the thalamic regions and the ACC (Fig. 3). The differences between PET and fMRI results can be explained by higher time resolution of fMRI, reflecting more dynamic changes.
Thalamic neurons change their firing mode from tonic to phasic, and as sleep deepens dorsal thalamic neurons get more hyperpolarized because of prolonged burst firing patterns of GABAergic reticular neurons (Steriade, 2001b). The resulting rhythmic activity then spreads into the cortex (Steriade, 2000), where additional inherent slow rhythmic activity is generated during NREM sleep. We noticed no significant differential decrease in thalamic activation with increasing sleep depth. On the other hand, cortical deactivation increased upon sleep deepening. This highlights the autonomous contribution of purely cortical-generated delta waves within the cortex, leading to a BOLD signal decrease due to neuronal hyperpolarization, as discussed in detail previously (Czisch et al., 2004).
There is some debate about whether the prefrontal cortex is of particular importance in sleep (Muzur et al., 2002; Kalinchuk et al., 2003). Delta EEG activity during SWS is most intense in fronto-central areas such as the PFC, and most positively correlated with the length of prior wakefulness, thus being associated with postulated ‘recovery processes’ during sleep (Horne, 1993). Our results show a predominantly fronto-central down-regulation of neuronal activity, but association cortices in the brain also reduce their activity incrementally from wakefulness to S2, which is further enhanced for most of them in SWS (Fig. 1). Several studies compared SWS with an alert awake state using either [18F]fluorodeoxyglucose-PET (Buchsbaum et al., 1989; Maquet et al., 1990) or H152O-PET (Braun et al., 1997; Hofle et al., 1997; Maquet et al., 1997; Kajimura et al., 1999); their findings are comparable to ours. During SWS we noted a decrease in activity in several cortical areas such as regions of the frontal cortex, the IPL, the cingulate gyrus, the right STG, the precuneus, the cuneus as well as regions of the basal ganglia. During SWS oscillations the cortex shows periodical and rich spontaneous activity which presumably represents the preserved capacity to process internally generated signals (Steriade, 2001a). Therefore, although large areas of the cerebral cortex show less BOLD activity during sleep, one may not conclude that the processing of information is fully cut-off, as the BOLD contrast between wakefulness and sleep does not allow for an absolute measure of neuronal activity.
In agreement with findings concerning insular activity (Critchley et al., 2004) and its relation to (interoceptive) awareness, the activity of the insula during the light sleep period (especially during S2 in the right hemisphere) decreases as consciousness does. The activity of the posterior cingulate gyrus was also reduced during all sleep stages (again more pronounced during S1), as most parts of the cingulate cortex were. In the hypotheses of a default mode of brain function (Raichle et al., 2001; Greicius et al., 2003) it is assumed that the ventral ACC and the posterior cingulate cortex (PCC) subserve a specific network for the default mode, i.e. resting state, of brain function with greater activity during resting states than during cognitive tasks. The activation of the areas is explained by continuously higher levels of alertness in expectation of environmental stimuli that are only reduced when focusing attention within the context of a specific task. We demonstrate a general reduction in activity reflecting fading alertness, with the PCC already dampened in S1, while ventral anterior cingulate cortex activity was only reduced in SWS, revealing a sleep stage specific grading in loss of alertness.
Finally, we observed brain regions (albeit small clusters) that showed increased BOLD-related activity during SWS when compared with wakefulness, mainly in the IFG, temporal areas, the parahippocampal gyrus and the cerebellum. It should be considered that the pattern of neuronal activation and deactivation during NREM sleep is influenced by pre-sleep activity patterns of the brain, thereby adding between-subject variance in our study. Therefore, one should be cautious about proposing an active role of the observed small brain regions with increased activity during sleep. Nevertheless, in a so-called transfer model it is assumed that memory consolidation depends on hippocampally initiated reinstantiation during SWS of distributed cortical activity patterns. These patterns characterize previous active behavioural states (Kali and Dayan, 2004). Whether the parahippocampal activation in our study during SWS is related to memory consolidation processes or not has to be addressed in future studies with experimentally controlled pre-sleep learning periods.
Hypothalamic connectivity during NREM sleep
The hypothalamus is of central importance for sleep regulation involved in a reciprocal network of wake-promoting nuclei in the brainstem as well as the lateral hypothalamus itself and sleep-promoting neurons inside the hypothalamic VLPO. Neuronal firing has been shown to be reduced during NREM sleep in the tuberomammillary nucleus, locus coeruleus, pons and dorsal raphe as a consequence of the increased activity of the inhibitory VLPO and the innervation of the activating lateral hypothalamus (Saper et al., 2001). In our study, due to limited spatial resolution of the data, we cannot differentiate hypothalamic subregions, or other structures like areas in the midbrain or pons important for sleep–wake regulation, serving these functions. Although we measured a relatively diffuse hypothalamic region we were able to identify a temporally correlated network throughout the brain (Table 5 and Fig. 2) which supposedly summarizes both wake- and sleep-promoting functional alterations in the cortical network activity. We observed a decreased activity of hypothalamic regions throughout the NREM sleep stages in accordance with a dominant contribution to the BOLD signal arising from the reduced activity of wake-promoting neurons. It was only during NREM sleep stages, rather than wakefulness, that some pronounced synchronous BOLD activity changes that correlated with the hypothalamus occurred: limbic structures, regions of the frontal and parietal cortex, the basal forebrain and the brainstem showed a similar time course of activation as hypothalamic regions did. This pattern of connectivity resembles the pathway of the ascending arousal system which sends projections from the brainstem and posterior hypothalamus throughout the forebrain (Saper et al., 2001), thereby modulating the system's arousal state. Again, areas described by Raichle et al. (2001) to be intrinsically activated during alert wakefulness (PCC) and medial prefrontal cortex (MPFC) show synchronicity with hypothalamic activity change throughout NREM sleep. The PCC is known to be tonically active during wakefulness and was described as a region gathering information about the external environment. The ventral MPFC consists of discrete areas and receives input concerning a huge range of sensory information. It is connected with limbic structures like the amygdalae, the hypothalamus, the midbrain and the brainstem. The increased connectivity of these areas may thus reflect a correlate of the up- and down-regulation of these areas of vigilance control during falling and staying asleep, as well as the differential fading of residual attention to the environment.
One limitation of our study is that the awake state of our subjects could be interpreted as relaxed dozing just before sleep onset and therefore did not represent an alert waking state. Furthermore, in order to facilitate falling asleep thalamic activity might have been reduced to isolate the system from external stimulus satiation such as background scanner noise. Consequentially, thalamic activity during S1 was markedly lowered. Compared with S1 the subsequent activity of S2 and SWS rose, but was sufficient to maintain sleep. The additional decrease in activity during S2 in the secondary auditory cortex (STG) and post-central gyrus could be interpreted in the context of auditory stimulation studies. These experiments revealed that cortical deactivation during sleep is most prominently associated with stimulation-induced increased EEG hyperpolarization during S2, whereas deep SWS lacks further BOLD signal decreases upon external stimulation (Czisch et al., 2004). Rare or deviant tones induce event related potential (ERP) responses in NREM sleep, but repetitive and continuous sound presentation elicits a rapid habituation as seen in EEG/ERP recordings, pointing towards minimal cortical processing (Caekebeke et al., 1990). While in our previous study acoustic stimuli were applied on top of the background fMRI sound and thus induced short deviant auditory experiences, the present study involved a continuous fMRI scanner noise. One might argue that the continuous low level background noise in the present experiment certainly leads to similar habituation with potentially reduced neuronal responses and less activation of acoustic cortex as described during wakefulness (Moelker and Pattynama, 2003). Habituation effects, supported by the passive noise protection, and adaptation sessions constituted a necessary prerequisite for falling and staying asleep. In our study subjects underwent 36 h of total sleep deprivation prior to the fMRI session. This protocol was chosen to compensate for increased sleep onset times in the uncomfortable MR environment and to minimize spontaneous or noise induced microarousals (Sforza et al., 2004). Similar sleep deprivation is regularly applied in H152O-PET studies (Braun et al., 1997; Hofle et al., 1997; Kajimura et al., 1999; Finelli et al., 2000; Balkin et al., 2002), but not in [18F]fluorodeoxyglucose-PET studies due to the longer time scales for uptake and decay of the radioactive tracer (Maquet et al., 1990, 1992; Nofzinger et al., 2002). Sleep deprivation may confound neuroimaging results by altering the sleep pattern in the recovery night, especially in the first sleep cycle as assessed in our study. It is known that the amount of SWS is doubled, and the latency to fall asleep as well as S1 and S2 are shortened (Dijk et al., 1993; Curcio et al., 2003). But sleep deprivation does not change the overall regional pattern of distinct EEG frequency bands during recovery sleep, leading to the assumption that the cerebral localization of sleep specific EEG components must be more closely related to the underlying generating neural mechanisms than to the level of sleep propensity (Finelli et al., 2001). Although it is unlikely that sleep deprivation leads to false activations in brain areas that might normally not alter their activity pattern during the sleep–wake cycle, our results might be slightly compromised by stronger delta activity due to higher statistical power.
Finally, a more detailed dynamic model of cortical activation during sleep could be assessed in principle using event-related fMRI with brief electrophysiological events more directly linked to BOLD responses or by defining more categories (epochs) at a smaller time scale (Ogilvie, 2001).
In summary, our study revealed several sleep stage specific brain regions showing less BOLD-related activity compared with wakefulness. This suggests that only if these regions interact in a well-balanced way a synchronized sleeping state can be established. One of these brain structures, the hypothalamic region, shows a specific hypothalamic-cortical pattern of connectivity only while the system is in the sleeping state. This underlines the role of hypothalamic-driven network regulation for initiation and maintenance of healthy human sleep.
The authors are grateful to Rosa Hemauer for her invaluable support and thank Armin Mann for the technical assistance.
References
Andersson JL, Onoe H, Hetta J, Lidstrom K, Valind S, Lilja A, et al. Brain networks affected by synchronized sleep visualized by positron emission tomography.
Balkin TJ, Braun AR, Wesensten NJ, Jeffries K, Varga M, Baldwin P, et al. The process of awakening: a PET study of regional brain activity patterns mediating the re-establishment of alertness and consciousness.
Braun AR, Balkin TJ, Wesenten NJ, Carson RE, Varga M, Baldwin P, et al. Regional cerebral blood flow throughout the sleep-wake cycle. An H2(15)O PET study.
Braun AR, Balkin TJ, Wesensten NJ, Gwadry F, Carson RE, Varga M, et al. Dissociated pattern of activity in visual cortices and their projections during human rapid eye movement sleep.
Brett M, Johnsrude IS, Owen AM. The problem of functional localization in the human brain.
Buchsbaum MS, Gillin JC, Wu J, Hazlett E, Sicotte N, Dupont RM, et al. Regional cerebral glucose metabolic rate in human sleep assessed by positron emission tomography.
Caekebeke JF, Van Dijk JG, Van Sweden B. Habituation of K-complexes or event-related potentials during sleep.
Creutzfeld OD. Cortex cerebri. Performance, structural and functional organization of the cortex. New York: Oxford University Press;
Critchley HD, Wiens S, Rotshtein P, Ohman A, Dolan RJ. Neural systems supporting interoceptive awareness.
Curcio G, Ferrara M, Pellicciari MC, Cristiani R, De Gennaro L. Effect of total sleep deprivation on the landmarks of stage 2 sleep.
Czisch M, Wetter TC, Kaufmann C, Pollmächer T, Holsboer F, Auer DP. Altered processing of acoustic stimuli during sleep: reduced auditory activation and visual deactivation detected by a combined fMRI/EEG study.
Czisch M, Wehrle R, Kaufmann C, Wetter TC, Holsboer F, Pollmächer T, et al. Functional MRI during sleep: BOLD signal decreases and their electrophysiological correlates.
Dement W, Kleitman N. Cyclic variations in EEG during sleep and their relation to eye movements, body motility, and dreaming.
Dijk DJ, Hayes B, Czeisler CA. Dynamics of electroencephalographic sleep spindles and slow wave activity in men: effect of sleep deprivation.
Drummond SP, Smith MT, Orff HJ, Chengazi V, Perlis ML. Functional imaging of the sleeping brain: review of findings and implications for the study of insomnia.
Ferrara M, De Gennaro L, Curcio G, Cristiani R, Bertini M. Regional differences of the temporal EEG dynamics during the first 30 min of human sleep.
Finelli LA, Landolt HP, Buck A, Roth C, Berthold T, Borbely AA, et al. Functional neuroanatomy of human sleep states after zolpidem and placebo: a H2(15)O-PET study.
Finelli LA, Borbely AA, Achermann P. Functional topography of the human nonREM sleep electroencephalogram.
Friston KJ, Holmes AP, Worsley KJ, Poline JB, Frith CD, Frackowiak RSJ. Statistical parametric maps in functional imaging: a general linear approach.
Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis.
Horne JA. Human sleep, sleep loss and behaviour. Implications for the prefrontal cortex and psychiatric disorder.
Hoffmann A, Jager L, Werhahn KJ, Jaschke M, Noachtar S, Reiser M. Electroencephalography during functional echo-planar imaging: detection of epileptic spikes using post-processing methods.
Hofle N, Paus T, Reutens D, Fiset P, Gotman J, Evans AC, et al. Regional cerebral blood flow changes as a function of delta and spindle activity during slow wave sleep in humans.
Kajimura N, Uchiyama M, Takayama Y, Uchida S, Uema T, Kato M, et al. Activity of midbrain reticular formation and neocortex during the progression of human non-rapid eye movement sleep.
Kali S, Dayan P. Off-line replay maintains declarative memories in a model of hippocampal-neocortical interactions.
Kalinchuk AV, Urrila AS, Alanko L, Heiskanen S, Wigren HK, Suomela M, et al. Local energy depletion in the basal forebrain increases sleep.
Kjaer TW, Law I, Wiltschiotz G, Paulson OB, Madsen PL. Regional cerebral blood flow during light sleep–a H2(15)O-PET study.
Krueger JM, Obal JF, Kapas L, Fang J. Brain organization and sleep function.
Lancaster JL, Woldorff MG, Parsons LM, Liotti M, Freitas CS, Rainey L, et al. Automated Talairach atlas labels for functional brain mapping.
Løvblad KO, Thomas R, Jakob PM, Scammell T, Bassetti C, Griswold M, et al. Silent functional magnetic resonance imaging demonstrates focal activation in rapid eye movement sleep.
Macey PM, Macey KE, Kumar R, Harper RM. A method for removal of global effects from fMRI time series.
Madsen PL, Holm S, Vorstrup S, Friberg L, Lassen NA, Wildschiodtz G. Human regional cerebral blood flow during rapid-eye-movement sleep.
Madsen PL, Schmidt JF, Holm S, Vorstrup S, Lassen NA, Wildschiodtz G. Cerebral oxygen metabolism and cerebral blood flow in man during light sleep (stage 2).
Madsen PL, Schmidt JF, Wildschiodtz G, Friberg L, Holm S, Vorstrup S, et al. Cerebral O2 metabolism and cerebral blood flow in humans during deep and rapid-eye-movement sleep.
Maquet P, Dive D, Salmon E, Sadzot B, Franco G, Poirrier R, et al. Cerebral glucose utilization during sleep-wake cycle in man determined by positron emission tomography and [18F]2-fluoro-2-deoxy-D-glucose method.
Maquet P, Dive D, Salmon E, Sadzot B, Franco G, Poirrier R, et al. Cerebral glucose utilization during stage 2 sleep in man.
Maquet P, Peters J, Aerts J, Delfiore G, Degueldre C, Luxen A, et al. Functional neuroanatomy of human rapid-eye-movement sleep and dreaming.
Maquet P, Degueldre C, Delfiore G, Aerts J, Peters JM, Luxen A, et al. Functional neuroanatomy of human slow wave sleep.
Moelker A, Pattynama PM. Acoustic noise concerns in functional magnetic resonance imaging.
Muzur A, Pace-Schott EF, Hobson JA. The prefrontal cortex in sleep.
Nofzinger EA, Mintun MA, Wiseman M, Kupfer DJ, Moore RY. Forebrain activation in REM sleep: an FDG PET study.
Nofzinger EA, Buysse DJ, Miewald JM, Meltzer CC, Price JC, Sembrat RC, et al. Human regional cerebral glucose metabolism during non-rapid eye movement sleep in relation to waking.
Peigneux P, Laureys S, Fuchs S, Delbeuck X, Degueldre C, Aerts J, et al. Generation of rapid eye movements during paradoxical sleep in humans.
Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function.
Rechtschaffen A, Kales A. A manual of standardized terminology, techniques and scoring system for sleep stages of human subjects. Washington DC: US Public Health Service, US Government Printing Office;
Saper CB, Chou TC, Scammell TE. The sleep switch: hypothalamic control of sleep and wakefulness.
Sforza E, Chapotot F, Pigeau R, Paul PN, Buguet A. Effects of sleep deprivation on spontaneous arousals in humans.
Steriade M. Corticothalamic resonance, states of vigilance and mentation.
Steriade M. Impact of network activities on neuronal properties in corticothalamic systems.
Author notes
1Max Planck Institute of Psychiatry, Munich, 2Centre of Mental Health, Klinikum Ingolstadt, Ingolstadt, 3Department of Psychology, Humboldt-Universität zu Berlin, Berlin, Germany and 4Academic Radiology, University of Nottingham, Queen's Medical Centre, Nottingham, UK