Abstract
The measurement of gene expression in postmortem brain is an important tool for understanding the pathogenesis of serious psychiatric disorders. We hypothesized that major molecular deficits associated with psychiatric disease would affect the entire brain, and such deficits may be shared across disorders. We performed RNA sequencing and quantified gene expression in the hippocampus of 100 brains in the Stanley Array Collection followed by replication in the orbitofrontal cortex of 57 brains in the Stanley Neuropathology Consortium. We then identified genes and canonical pathway gene sets with significantly altered expression in schizophrenia and bipolar disorder in the hippocampus and in schizophrenia, bipolar disorder and major depression in the orbitofrontal cortex. Although expression of individual genes varied, gene sets were significantly enriched in both of the brain regions, and many of these were consistent across diagnostic groups. Further examination of core gene sets with consistently increased or decreased expression in both of the brain regions and across target disorders revealed that ribosomal genes are overexpressed while genes involved in neuronal processes, GABAergic signaling, endocytosis and antigen processing have predominantly decreased expression in affected individuals compared to controls without a psychiatric disorder. Our results highlight pathways of central importance to psychiatric health and emphasize messenger RNA processing and protein synthesis as potential therapeutic targets for all three of the disorders.
Similar content being viewed by others
Introduction
Schizophrenia (SCZ), bipolar disorder (BPD) and major depression (MDD) are complex neuropsychiatric disorders with etio-pathogenic factors that likely include both genetic and environmental components. Genome-wide association studies (GWAS) have identified polymorphisms that contribute to the risk of developing SCZ, BPD or MDD.1, 2, 3, 4, 5 SCZ and BPD are spectrum disorders with overlapping symptoms, suggesting that there is biological overlap between them. In fact, the most informative polymorphisms are similarly predictive of SCZ, BPD and MDD with correlated effects sizes for each disorder.1, 5 Most of the polymorphisms that associate with psychiatric disorders are outside of protein coding regions and likely alter gene expression rather than protein structure.1, 3 Functional analysis of genetic variants that associate with SCZ, BPD and MDD found that pathways that regulate histone methylation and thereby gene transcription are the most strongly implicated on the basis of the shared genetics across the three disorders, followed by immune and signaling pathways.3 However, the downstream impact of these polymorphisms on gene expression in the brain remains unclear. Gene set expression studies have reproducibly implicated a few key functions in particular disorders, but the core processes that are common across diagnostic groups and brain regions are not well established. Most expression studies of functionally related gene sets in postmortem brains have focused on a single brain region6, 7, 8, 9, 10, 11, 12, 13 and/or a single disorder.6, 7, 9, 13
Deficits associated with psychiatric disease have been discovered in nearly every brain region that has been studied. Compared with healthy controls, the volume of various brain regions including hippocampus, amygdala, thalamus, nucleus accumbens, pallidum, lateral ventricles14 and the supragranular cortical layers15 are reported to be different in SCZ. Gray and white matter deficits in SCZ have also been reported in the superior temporal gyrus, left medial temporal lobe,16 frontal temporal lobe, occipital lobe17 and other regions.18 Individuals with SCZ also have higher GABA neuron density in the white matter of the orbitofrontal cortex (OFC) in SCZ,19 and decreased connectivity between the cerebellum and the thalamus and the frontal cortex.20 Functional deficits in SCZ include impaired functioning of the magnocellular visual pathway,21 altered contextual modulation of critical responses in the primary visual cortex.22 Anatomical differences in the left and right superior temporal cortex, left supramarginal/angular gyrus, left postcentral gyrus and left posterior cingulate cortex correlate with the severity of auditory hallucinations.23 In addition, decreased cerebellar24 and hippocampal volume25 and anatomical differences in the gray mater of the frontal temporal lobe17 have been reported in BPD, and reduced gray matter volume in the OFC has been reported in MDD.26 Accordingly, previous studies have identified differences in gene or protein expression in the lateral cerebella, frontal cortex, prefrontal cortex, OFC, dorsolateral prefrontal cortex, anterior cingulate cortex, superior temporal gyrus and hippocampus in SCZ, BPD and/or MDD.7, 8, 9, 10, 11, 12, 13, 27, 28, 29, 30, 31, 32, 33, 34, 35 These findings suggest that molecular deficits underlying psychiatric disease are not restricted to a particular region and likely affect the entire brain.
In particular, genes that are dysregulated as a result of genetic variants are likely to be affected throughout the brain as the variants are present in every cell. On the basis of this rationale, several large expression and epigenetic studies have been conducted in peripheral blood lymphocytes.36, 37, 38 A comparison of the co-expression of genes in human brain and human blood revealed co-expression modules that are preserved across tissues, supporting the notion that polymorphisms will result in similar deficits in different cell types.39 Furthermore, the expression levels of key genes in the preserved modules tended to be more heritable than genes in modules that were not preserved. Therefore, it seems likely that mechanisms that regulate gene expression could affect multiple brain regions in the same way. We further reasoned that although not all changes in gene expression will be consistent across brain regions, those that are consistent are more likely to represent core pathways underlying the etiology of these complex disorders. We therefore examined gene expression in the hippocampus, which is part of the limbic system involved in learning and memory, and replicated these findings in a separate cohort using samples from the OFC, which has been implicated in mood disorders.17, 26 Our first cohort consisted of 100 hippocampal samples from the Stanley Array Collection, which includes individuals with SCZ, BPD and controls without a psychiatric disorder. Our second cohort consisted of 57 OFC samples from the Stanley Neuropathology Consortium, which includes individuals with SCZ, BPD, MDD and controls. For each diagnostic group, we identified expression differences relative to matched controls and ranked all genes according to the strength and direction of differential expression. Using the gene set enrichment analysis (GSEA) tool,40 which scores sets of genes according to their distribution within a ranked list, we identified gene sets containing a disproportionate number of genes with either increased or decreased expression in each diagnostic group. We identified gene sets that replicated across brain regions in each disorder and found that many of the replicated gene sets were consistently perturbed in all three disorders. We further examined 13 universal gene sets that were enriched in the same direction in both the hippocampus and the OFC in SCZ, BPD and MDD.
Materials and methods
Postmortem brain samples and RNA sequencing
We performed ultra-high-depth strand-specific RNA-seq using RNA isolated from 102 hippocampal samples in the Stanley Array Collection and a replication cohort of 59 OFC samples in the Stanley Neuropathology Consortium.41 Libraries were prepared and sequenced as detailed in the Supplementary Information. To minimize technical batch effects, each library was run on a full sequencing lane and samples from all diagnostic groups were distributed randomly across the sequencing runs. All of the raw sequencing files and topHat alignments are available for download at www.sncid.org. Two outliers were removed from each collection on the basis of the results of principal components analyses detailed in the supplement and Supplementary Figure S1. The remaining samples are well matched for demographic factors including age, sex, race, postmortem interval (PMI), pH and the side of the brain used to extract RNA (Table 1).
Gene expression analysis
The details are presented in the Supplementary Information. In brief, we aligned sequencing reads to the human genome (hg19) using the TopHat2 short read aligner42 and counted the reads aligning to exons in each gene annotated in the UCSC Genome Bioinformatics hg19 knownGene table. We then used DESeq2 (ref. 43) to normalize raw read counts and identify differences in gene expression between each diagnostic group and matched controls using age, sex, brain pH and PMI as factors in the design formula and diagnosis as the condition of interest.
Gene set enrichment and functional network analysis
The details are presented in the Supplementary Information. For each diagnostic group, we ranked all genes according to the Wald test statistic for differential expression provided by DESeq2.43 Using the GseaPreranked tool in GSEA,40 we identified gene sets that were enriched among overexpressed or under-expressed genes with a false discovery rate (FDR) q<0.05. We utilized the EnrichmentMap44 application for Cytoscape45, 46 to map the enriched gene sets into functional networks.
Results
Differential expression in the hippocampus
We sequenced 100 hippocampal samples to produce an average of 154 000 000 paired-end 100 base reads that aligned to the human genome (hg19; Supplementary Table S1). After counting the reads aligning to each gene, we used DESeq2 to compare gene expression in each diagnostic group with matched controls. We accounted for demographic variables in the differential expression analysis using a multi-factor design formula that includes sex, age, PMI and pH as covariates and diagnosis as the contrast variable. We detected expression from 21 861 genes in the hippocampus, most of which were not differentially expressed after multiple testing correction (Supplementary Figure S2). We found 12 genes that were differentially expressed in SCZ and seven in BPD (Table 2). No genes were differentially expressed in both SCZ and BPD.
To identify concerted changes in gene expression that accompany psychiatric disorders, we used GSEA40 to identify canonical pathway gene sets containing a disproportionate number of genes with either increased or decreased expression. For each disorder, we ranked all the genes using the Wald test statistic from DESeq2, which correlates with the amplitude and significance of expression differences, then identified gene sets that are enriched among overexpressed or under-expressed genes. We identified 89 gene sets enriched in SCZ and 170 in BPD (Supplementary Tables S2 and S3) with FDR q-values below 0.05.
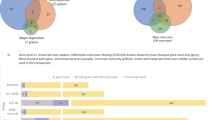
Because we used gene sets from all major gene ontology databases, we expected many sets to be comprised of identical or highly overlapping groups of genes. We therefore utilized the EnrichmentMap application in Cytoscape44, 45, 46 to analyze the proportion and identity of genes from our expression data sets that are in common across the enriched gene sets (Supplementary Tables S4 and S5) and to visualize the networks created by overlapping gene sets (Figure 1, enlarged in Supplementary Figures S3 and S4). In the enrichment maps, each significant gene set from GSEA is represented by a circle. Overlapping gene sets are connected into networks by lines that represent shared genes. The size of the circles represents the number of genes in our expression data sets that are annotated in each gene set, while the thickness of the lines represents the proportion of genes that are in common between gene sets. On the basis of the number and thickness of interconnected lines, most of the gene sets that were enriched in GSEA had genes in common; they formed highly interconnected clusters describing interrelated functions. In SCZ, the largest network indicates upregulation of genes involved in messenger RNA (mRNA) processing and translation, some of which also contain genes involved in transport of ribonucleoproteins, which in turn shares genes related to other types of transmembrane transport, interferon signaling and cytokine signaling (Figure 1a, Supplementary Figure S3). In BPD, genes involved in mRNA processing and translation make up the second largest cluster. The largest cluster is of 28 highly overlapping downregulated gene sets involved in major histocompatibility (MHC) Class I antigen presentation, ubiquitin-mediated degradation by the proteasome and related signaling networks (Figure 1b, Supplementary Figure S4).
Functional analysis of overlap between sets of genes enriched in SCZ (a) and BPD (b) in the hippocampus. Red circles denote gene sets with increased expression-affected individuals, whereas blue circles indicate decreased expression. The size of each circle represents the number of genes in each set, while the thickness of the lines connecting two circles represents the proportion of genes that are in common between the two gene sets. A fully labeled version of each figure is available in the Supplementary Information (Supplementary Figures S2 and S3). The major functional networks are as follows: (a1) messenger RNA (mRNA) processing and translation, transport of ribonucleoproteins, transmembrane transport, interferon signaling and cytokine signaling; (a2) respiration/oxidative phosphorylation and genes implicated in Parkinson’s, Alzheimer’s and Huntington’s diseases; (a3) developmental biology, transcriptional regulation and axon guidance; (a4) defensins, innate immunity and complement cascade; (a5) neuronal systems and neural transmitter signaling; (a6) extracellular matrix proteins; (a7) endocytosis and intracellular trafficking; (b1) cell cycle regulation; (b2) Major histocompatibility (MHC) Class I antigen presentation, ubiquitin-mediated degradation by the proteasome and related signaling networks; (b3) mRNA processing and translation; (b4) protein folding and import into mitochondria; (b5) transcription termination, mRNA splicing and transport, interferon and cytokine signaling; (b6) neuronal signaling cascades; (b7) respiration/oxidative phosphorylation and genes implicated in Parkinson’s, Alzheimer’s and Huntington’s diseases; (b8) phospholipid metabolism; (b9) fatty acid and keytone metabolism, and regulation of peroxisomes and adipocites; (b10) transfer RNA (tRNA) biosynthesis; (b11) RNA polymerase III regulation; (b12) biological oxidations and drug metabolism by cytochrome p450. BPD, bipolar disorder; SCZ, schizophrenia.
Replication in the OFC
As we hypothesized that major molecular deficits underlying psychiatric disease would likely affect the entire brain, we performed a replication study using OFC samples from a separate cohort. We sequenced 57 OFC samples to produce an average of 140 000 000 single-end 100 base reads that aligned to the human genome (Supplementary Table S2). As with the hippocampus, we counted reads aligning to each gene and compared gene expression in each diagnostic group to matched controls using a multi-factor design that includes sex, age, PMI and pH as covariates. We detected expression from 20 711 genes in the OFC, most of which were not differentially expressed after multiple testing correction (Supplementary Figure S2). No genes were differentially expressed in SCZ, but 246 genes were significant in BPD (Supplementary Table S7) and 26 in MDD compared with controls (Supplementary Table S8).
As expected on the basis of the results of previous work,30 none of the individual genes that were differentially expressed in the hippocampus were replicated in the OFC. This may be, in part, because several of the genes that are differentially expressed in the hippocampus have low average read counts in the OFC (Supplementary Table S9), indicating that expression of these genes may be specific to each brain region. However, SCL11A1, MT1X, S100A8 and NR4A2 were differentially expressed in SCZ in the hippocampus and nominally significant in SCZ in the OFC. Also, FKBP5 and MT1X were nominally significant in BPD in the hippocampus and significant in BPD in the OFC (Supplementary Table S9).
In contrast to individual genes, there was greater concordance between the two brain regions when expression was examined in the context of functionally related gene sets. GSEA revealed 216 gene sets that were enriched in SCZ, 491 in BPD and 257 in MDD (Supplementary Tables S10–S12) with FDR q-values below 0.05. As with the gene sets enriched in the hippocampus, the significant gene sets overlapped extensively to form dense interconnected networks of genes with related functions (Supplementary Tables S13–S15, Supplementary Figures S5–S7). Twenty (22.5%) gene sets enriched in SCZ and 67 (39.4%) enriched in BPD in the hippocampus were also enriched in the same direction in the same disorder in the OFC. Although some replicated gene sets were enriched only in one disorder (Supplementary Table S16), most of the gene sets that replicated across brain regions in BPD were also enriched in MDD (Table 3, Supplementary Table S17). Thirteen gene sets replicated across brain regions in both SCZ and BPD, and all of these were also enriched in MDD in the OFC (Table 3). In addition to the 13 universal gene sets, the ‘Kyoto Encyclopedia of Genes and Genomes (KEGG) Calcium Signaling’ set was also replicated in both SCZ and BPD and enriched in MDD, but had decreased expression in BPD and MDD and increased expression in SCZ (Supplementary Table S16 and S17). Gene sets that are shared across diagnostic groups are less likely to be affected by medication use, which varied between diagnostic groups (Table 1). Although nearly all individuals with SCZ received antipsychotics, fewer individuals with BPD received antipsychotics and generally at lower dosages. No one in the MDD or control groups took antipsychotics. Conversely, most MDD patients used antidepressants, whereas antidepressant usage was less prevalent in the BPD and SCZ groups.
We plotted the overlap of the genes in the 13 universal gene sets (Supplementary Figure S8, Supplementary Tables S18–S22) and found that they actually denote four distinct groups. The strongest enrichment scores, particularly in BPD and MDD, were for nine gene sets involved in mRNA processing and translation (Table 3). Together, the nine overlapping gene sets contain a total of 338 genes from our expression data sets, including a core set of 80 genes that are in all the nine sets and comprise most of the ‘KEGG Ribosome’ set. Expression was higher in BPD than control in 78 of the 80 genes in hippocampus and in 79 genes in the OFC. Expression was also higher in 78 of the genes in MDD. In SCZ, expression was higher in 56 genes in the hippocampus and in 52 in the OFC. The high degree of overlap (Supplementary Figure S8) suggests that a core group of genes could drive enrichment of all nine functionally related sets.
To identify the genes that drive enrichment of each universal gene set, we performed a leading edge analysis in GSEA40 (Supplementary Tables S18–S23, Figure 2). Many of the same genes drive enrichment of all of the ribosomal gene sets. However, additional genes that are not part of the KEGG Ribosome set also drive enrichment of related sets with specialized functions. Supplementary Tables S18–S22 list the genes in each analysis that drive enrichment of the universal gene sets. Depending on the diagnostic group and brain region, between 135 and 218 genes drive expression of at least one of the ribosomal gene sets. Of these, between 49 and 70 (26–52% of the related genes in each analysis) are present in all the nine sets, whereas between 27 and 60% drive enrichment of only one set (Supplementary Table S23). In contrast, there is very little overlap between the genes driving enrichment of the four sets with decreased expression in BPD, MDD and SCZ (Figure 2). In each of the five analyses, between 333 and 366 genes drive enrichment of the four sets, with no genes driving enrichment of all four sets and 17 to 22 genes driving more than one set (Supplementary Tables S18–S23). The most strongly downregulated are neuronal system genes, followed by genes involved in antigen processing, GABA signaling and endocytosis (Table 3).
Overlap between lists of genes driving enrichment of gene sets. Darker green indicates higher degree of overlap. As different genes may contribute to enrichment of the gene sets in each sample group, separate heat map is plotted for each comparison. The top two plots denote genes with increased expression in SCZ and BPD relative to control in the hippocampus, while the bottom three plots denote genes with increased expression in SCZ, BPD and MDD in the OFC. BPD, bipolar disorder; MDD, major depressive disorder; mRNA, messenger RNA; OFC, orbitofrontal cortex; SCZ, schizophrenia; UTR, untranslated region.
In addition to the 13 universal gene sets, 35 sets were enriched in all mood disorder samples but not SCZ (Supplementary Table S17) including genes involved in synaptic signaling, translation, opioid signaling, transfer RNA synthesis and MHC class II antigen presentation. Because of the large number of gene sets that are enriched in all mood disorder samples, we further examined the overlap between BPD and MDD in the OFC. Although only two of the 26 genes that were differentially expressed in MDD were also significant in BPD, 182 (71%) of the 257 gene sets that are enriched in MDD were enriched in the same direction in BPD. As all the OFC samples were compared with the same controls, we also examined the overlap between BPD and SCZ in the OFC. Of 216 gene sets enriched in SCZ, 83 (38%) were enriched in the same direction in BPD.
In addition to the shared gene sets, 19 additional sets replicated in BPD but were not enriched in MDD (Supplementary Table S16). These include genes implicated in Alzheimer’s and Huntington’s diseases, as well as phosphatidylinositol signaling, aquaporin-mediated transport, adipocytokine signaling and regulation of the cell cycle. Six gene sets replicated in SCZ alone, including olfactory signaling, class I MHC-mediated antigen processing and membrane trafficking (Supplementary Table S16).
Discussion
We identified gene sets describing key biological processes with perturbed expression in SCZ and BPD in the postmortem hippocampus, then replicated our results in the OFC of a separate cohort. Our initial analysis included high-depth RNA-seq of 100 hippocampal samples from demographically well-matched individuals from the Stanley Array Collection and we replicated our findings in 57 OFC samples from the Stanley Neuropathology Consortium. We identified 13 gene sets that replicated across brain regions in SCZ and BPD, and were enriched MDD. The strongest enrichment scores were for nine sets describing functions related to mRNA processing, ribosome biogenesis and translation, all of which were overexpressed in SCZ, BPD and MDD. In addition, gene sets describing neuronal systems, antigen degradation, GABAergic signaling and endocytosis had decreased expression in all three of the psychiatric disorders. These gene sets describe processes previously implicated in SCZ, BPD or MDD by others using RNA-seq,6, 8, 9, 30, 47 microarrays6, 10, 11, 12, 48 or protein profiling49, 50 of postmortem brain samples. Our findings establish the central importance of these pathways by demonstrating that they are consistently altered in all diagnosis groups in two brain regions from two cohorts.
Our approach is aimed at identifying the core molecular deficits associated with psychiatric disease, so we added an additional level of stringency by using different brain regions from each cohort. The rationale for this approach is based on the fact that disease-associated genetic variants that regulate gene expression (that is, eQTLs) are present in every cell and therefore likely alter gene expression in multiple brain regions. Accordingly, altered gene expression in many brain regions has been reported in psychiatric disease.7, 8, 9, 10, 11, 12, 13, 27, 28, 29, 30, 31, 32, 33, 34, 35 Although we do not suggest that all changes in gene expression associated with disease will be consistent across brain regions, we provide evidence for the existence of robust changes in expression of certain gene pathways in multiple brain regions. Just as co-expression modules that are preserved across the brain and blood are driven by genes with more heritable gene expression levels than modules that are tissue specific,39 the changes in gene expression that are consistent across brain regions are more likely to represent core molecular deficits that are fundamental to disease development. Although our hypothesis is that the gene pathways we identified are likely disrupted in multiple brain regions, it is important to note that both the hippocampus and the OFC encode parallel but interactive cognitive maps.51 Potentially, the molecular deficits we describe might specifically disrupt such functions in the OFC and the hippocampus and thus contribute to the cognitive deficits observed in psychiatric disorders.
In addition to being present throughout the brain, many genetic variants implicated in psychiatric disease associate with multiple disorders.1, 5 The fact that we were able to identify gene pathways that are consistently dysregulated in different disorders as well as across brain regions and cohorts supports the hypothesis that a core set of molecular deficits may exist in psychiatric disorders. The limitation of this approach is that we may miss changes that are specific to a single brain region or disorder. Any such changes are likely included among the results presented in the supplement, but further replication studies in the hippocampus and OFC are needed to identify such differences.
Our results are consistent with findings from GWAS, which conclude that many genes of small effect size contribute to the pathology of psychiatric disorders. Similar to previous postmortem expression studies, we find subtle differences in the expression levels of individual genes between psychiatric cases and unaffected controls.8, 29, 30 Using gene set enrichment analysis, which measures small but consistent changes in expression of genes that work together in a biological pathway,40 we identified a number of gene pathways that appear to be consistently altered in psychiatric disorders. Each individual comparison, for example, SCZ vs CONTROL in hippocampus, yields many enriched gene sets but, based on our functional network analysis of the enriched gene sets, most describe related pathways and their enrichment is driven by an overlapping set of genes. The fact that expression of multiple gene pathways is affected in psychiatric disease is not surprising given that there are 108 different genetic regions associated with SCZ4 alone and a polygenic risk score is the most accurate predictor of psychiatric disease.52 In addition, disease-associated polymorphisms are enriched in genes involved in posttranslational modification of histones, a key regulator of transcription in the cell.3 Thus, our expression results are highly congruent with genetic findings and support the notion that many genes of small effect size contribute to the pathology of psychiatric disorders.
This study differs from previous RNA-seq studies8, 29, 30 that identified genes that are dysregulated in SCZ and BPD using different samples from the Stanley brain collections. We provide RNA sequencing results from brain regions that have not previously been sequenced and apply analysis methods designed to identify the overarching trends in gene expression that are in common across brain regions. Kim et al.8 and Zhao et al.30 both noted difficulty in replicating differential expression of individual genes. As both studies utilized hypergeometric tests for pathway enrichment based on significant genes, the lack of replication at the gene level proved problematic. Kim et al. therefore focused on gene–gene interactions revealed by co-expression analysis, whereas Zhao et al. selected the genes from the replication set with the greatest expression differences although none reached significance. We instead used expression information from all the genes to identify pathways enriched in each diagnostic group using GSEA. We then replicated the gene set enrichment in a different brain region from an additional cohort and identified validated pathways that are in common across multiple diagnostic groups.
We accounted for the effects of sex, age, brain pH and PMI by including these variables as covariates while determining differential expression of each gene. Although we were not able to specifically test the effect of medications on gene set enrichment, we accounted for antipsychotic use by evaluating gene expression in each diagnostic group independently and focusing on gene sets that were universally perturbed. All of the gene sets that replicated in both SCZ and BPD were also enriched in MDD although the MDD group did not use antipsychotics. Similarly, more individuals in the MDD group than in the BPD and SCZ groups took antidepressants. Further confidence that our results are not driven by medication use comes from a comparison of gene expression in postmortem brains10 in which subjects with BPD exposed to antipsychotics had gene expression levels that were closer to those of control individuals than medication-naive patients.
The nine gene sets overexpressed in SCZ, BPD and MDD all contain genes from the KEGG ribosome set. Each set also contains additional dysregulated genes, suggesting that multiple functional pathways involving ribosomes are affected in the three target disorders. Affected processes include ‘nonsense mediated decay enhanced by the exon junction complex’, ‘metabolism of mRNA’, ‘viral RNA transcription and replication’, ‘3′-UTR (untranslated region)-mediated translational regulation’, ‘peptide chain elongation’ and ‘translation’. Our results indicate that the magnitude and significance of overexpression of ribosomal genes is greater in mood disorder samples than SCZ, but is significant in all the three disorders. Increased expression of ribosomal genes in SCZ has also been noted in other recent studies. ‘Translational elongation’, ‘ribosome biogenesis’ and ‘ribosomal RNA processing’ gene sets were enriched in a co-expression module in the hippocampus with increased expression in SCZ,8 and the ‘ribosome’, ‘translation’, ‘translational elongation’ and the 'ribosomal subunit' GO terms were enriched among genes differentially expressed in SCZ in the superior temporal gyrus.13 Also, protein expression profiling of human induced pluripotent stem cell-derived neural progenitor cells revealed increased levels of ribosomal proteins and translation factors as well as increased translational activity and total protein levels in cells from SCZ patients relative to controls.53 In addition, co-expression modules associated with BPD in the dorsolateral prefrontal cortex that included both upregulated and downregulated genes were enriched for ‘RNA processing’ and ‘ribosomal subunit’ GO terms,9 and ribosomal genes were overexpressed in the hippocampus of mice modeling a Creb1 promoter mutation that causes familial MDD.54 Several forms of synaptic plasticity may require the activation of stalled polyribosomes that have initiated translation and are paused before peptide elongation.55, 56 Intriguingly, ketamine and other rapid-acting antidepressants work in part via phosphorylation cascades triggered by the mTOR pathway that ultimately stimulate translation of synaptogenic proteins,57 suggesting that translational deficits may exist in untreated individuals. It will, therefore, be interesting to study whether increased transcription of ribosomal protein genes in the hippocampus and OFC correlates with increased translation or with a buildup of stalled polyribosomes.
Many of the gene sets that we found to have decreased expression in SCZ, BPD and MDD also overlap with genes previously implicated in one or two disorders. The ‘Reactome Neuronal Systems’ gene set contains ‘synaptic proteins’ genes that are differentially co-regulated in BPD and MDD in the hippocampus8 and dysregulated in BPD in the premotor cortex.10 In the prefrontal cortex, genes involved in nervous system development,12 synaptic density and plasticity30 have altered expression in SCZ and BPD. In addition, genes that impact long-term potentiation have altered expression in the anterior cingulate cortex in SCZ.49 The role of GABAergic signaling in SCZ is also established,6, 8, 47, 58 consistent with our findings. Defective regulation of antigen processing and ubiquitination is supported by the finding that ubiquitin-mediated degradation genes are dysregulated in BPD48 and by protein expression differences in ubiquitination and degradation pathways in the hippocampus in SCZ and BPD.50 Dysregulation of ‘endocytosis’ genes is supported by the previous finding that proteins in the endocytosis pathway have altered expression in SCZ in the postsynaptic density49 and gene pathways such as ‘Fc gamma receptor-mediated phagocytosis’ are dysregulated in BPD and SCZ in the cingulate cortex.30 Together, ‘antigen processing via ubiquitination-mediated proteasome degradation’ to make class I MHC molecules and ‘endocytosis’ in the form of Fc gamma receptor-mediated phagocytosis contribute to the adaptive immune system, which has repeatedly been implicated in SCZ59, 60 and MDD.61 Markers of inflammation and innate immunity also frequently have altered expression in psychiatric disorders.6, 8, 12, 59, 60, 61 We found that gene sets related to innate immunological processes such as cytokine signaling were enriched in SCZ, BPD and MDD in both the OFC and the hippocampus, but the direction of the differential expression was not consistent between brain regions or diagnostic groups. Accordingly, while dysregulation of innate immunity has been established in both SCZ and BPD, expression of innate immunity genes represents a major difference between the diseases.8, 11
Our observation that key pathways are concordantly regulated in SCZ, BPD and MDD is consistent with GWAS results as polygenic risk scores for SCZ, BPD and MDD show some cross-disorder association1 and disease-associated loci suggest the involvement of immune and neuronal signaling in all the three disorders.3 In particular, the MHC locus, which encodes components of the MHC Class I and II complexes, is strongly associated with SCZ and BPD62 and key genes within the locus have been shown to have decreased expression in SCZ and BPD throughout the brain.63 Accordingly, the Reactome MHC Class II antigen presentation gene set had decreased expression in all mood disorder samples, whereas the Reactome Class I MHC-mediated antigen processing presentation gene set had decreased expression in SCZ. Calcium signaling has also been implicated in SCZ, BPD and MDD based on GWAS. We found that dysregulation of calcium signaling genes was reproducible in both SCZ and BPD, but expression was increased in SCZ and decreased in mood disorder samples. As genetic variation can affect gene expression in a variety of ways, our findings fit with the GWAS results and may indicate that some variants have a context-specific impact on gene expression.
Although no single gene was differentially expressed in the same disorder in both brain regions, MT1X, SLC11A1, S100A8 and NR4A2 were significant in SCZ in the hippocampus and nominally significant in the OFC. In addition, MT1X and FKBP5 reached nominal significance in BPD in the hippocampus and were significant in the OFC. MT1X encodes a metallothionein that may be neuroprotective during stress response and is dysregulated in postmortem brains in psychosis64 and suicide.65 SLC11A1 encodes an iron transporter with a key role in innate immunity.66 S100A8 encodes a calcium-binding protein that regulates inflammation and immune response and has altered expression in blood from SCZ patients.67 NR4A2 encodes a transcription factor regulating dopaminergic neurosynthesis68 previously shown by in situ hybridization and western blotting to have decreased expression in postmortem brains in psychiatric disease.69, 70 FKBP5 encodes a key regulator of glucocorticoid response, is widely implicated in psychiatric disease and was previously identified as overexpressed in the frontal cortex in SCZ, BPD and MDD.71, 72
We performed one of the largest RNA sequencing studies of postmortem brain tissue in psychiatric disease and discovered that specific gene sets are dysregulated across cohorts, brain regions and disorders. The fact that the gene sets identified in this study are consistently altered across brain regions suggests that they may represent fundamental molecular deficits associated with psychiatric disease. The further characterization of biological processes described by these gene sets has the potential to reveal important insights into disease pathology and may finally unravel the etiology of psychiatric disorders.
References
Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013; 381: 1371–1379.
Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry 2013; 18: 497–511.
Network, Pathway Analysis Subgroup of Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci 2015; 18: 199–209.
Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–427.
Schulze TG, Akula N, Breuer R, Steele J, Nalls MA, Singleton AB et al. Molecular genetic overlap in bipolar disorder, schizophrenia, and major depressive disorder. World J Biol Psychiatry 2014; 15: 200–208.
Hwang Y, Kim J, Shin JY, Kim JI, Seo JS, Webster MJ et al. Gene expression profiling by mRNA sequencing reveals increased expression of immune/inflammation-related genes in the hippocampus of individuals with schizophrenia. Transl Psychiatry 2012; 3: e321.
Cruceanu C, Tan PP, Rogic S, Lopez JP, Torres-Platas SG, Gigek CO et al. Transcriptome sequencing of the anterior cingulate in bipolar disorder: dysregulation of G protein-coupled receptors. Am J Psychiatry 2015; 172: 1131–1140.
Kim S, Hwang Y, Webster MJ, Lee D . Differential activation of immune/inflammatory response-related co-expression modules in the hippocampus across the major psychiatric disorders. Mol Psychiatry 2016; 21: 376–385.
Akula N, Wendland JR, Choi KH, McMahon FJ . An integrative genomic study implicates the post-synaptic density in the pathogenesis of bipolar disorder. Neuropsychopharmacology 2015; 41: 886–895.
Chen H, Wang N, Zhao X, Ross CA, O'Shea KS, McInnis MG . Gene expression alterations in bipolar disorder postmortem brains. Bipolar Disord 2013; 15: 177–187.
de Baumont A, Maschietto M, Lima L, Carraro DM, Olivieri EH, Fiorini A et al. Innate immune response is differentially dysregulated between bipolar disease and schizophrenia. Schizophr Res 2015; 161: 215–221.
Shao L, Vawter MP . Shared gene expression alterations in schizophrenia and bipolar disorder. Biol Psychiatry 2008; 64: 89–97.
Wu JQ, Wang X, Beveridge NJ, Tooney PA, Scott RJ, Carr VJ et al. Transcriptome sequencing revealed significant alteration of cortical promoter usage and splicing in schizophrenia. PLoS One 2012; 7: e36351.
van Erp TG, Hibar DP, Rasmussen JM, Glahn DC, Pearlson GD, Andreassen OA et al. Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol Psychiatry 2016; 21: 585.
Wagstyl K, Ronan L, Whitaker KJ, Goodyer IM, Roberts N, Crow TJ et al. Multiple markers of cortical morphology reveal evidence of supragranular thinning in schizophrenia. Transl Psychiatry 2016; 6: e780.
Honea R, Crow TJ, Passingham D, Mackay CE . Regional deficits in brain volume in schizophrenia: a meta-analysis of voxel-based morphometry studies. Am J Psychiatry 2005; 162: 2233–2245.
Anderson D, Ardekani BA, Burdick KE, Robinson DG, John M, Malhotra AK et al. Overlapping and distinct gray and white matter abnormalities in schizophrenia and bipolar I disorder. Bipolar Disord 2013; 15: 680–693.
Tang J, Liao Y, Zhou B, Tan C, Liu W, Wang D et al. Decrease in temporal gyrus gray matter volume in first-episode, early onset schizophrenia: an MRI study. PLoS One 2012; 7: e40247.
Joshi D, Fung SJ, Rothwell A, Weickert CS . Higher gamma-aminobutyric acid neuron density in the white matter of orbital frontal cortex in schizophrenia. Biol Psychiatry 2012; 72: 725–733.
Chen YL, Tu PC, Lee YC, Chen YS, Li CT, Su TP . Resting-state fMRI mapping of cerebellar functional dysconnections involving multiple large-scale networks in patients with schizophrenia. Schizophr Res 2013; 149: 26–34.
Martinez A, Hillyard SA, Dias EC, Hagler DJ Jr., Butler PD, Guilfoyle DN et al. Magnocellular pathway impairment in schizophrenia: evidence from functional magnetic resonance imaging. J Neurosci 2008; 28: 7492–7500.
Seymour K, Stein T, Sanders LL, Guggenmos M, Theophil I, Sterzer P . Altered contextual modulation of primary visual cortex responses in schizophrenia. Neuropsychopharmacology 2013; 38: 2607–2612.
Nenadic I, Smesny S, Schlosser RG, Sauer H, Gaser C . Auditory hallucinations and brain structure in schizophrenia: voxel-based morphometric study. Br J Psychiatry 2010; 196: 412–413.
Baldacara L, Nery-Fernandes F, Rocha M, Quarantini LC, Rocha GG, Guimaraes JL et al. Is cerebellar volume related to bipolar disorder? J Affect Disord 2011; 135: 305–309.
Chepenik LG, Wang F, Spencer L, Spann M, Kalmar JH, Womer F et al. Structure-function associations in hippocampus in bipolar disorder. Biol Psychol 2012; 90: 18–22.
Lacerda AL, Keshavan MS, Hardan AY, Yorbik O, Brambilla P, Sassi RB et al. Anatomic evaluation of the OFC in major depressive disorder. Biol Psychiatry 2004; 55: 353–358.
Akula N, Barb J, Jiang X, Wendland JR, Choi KH, Sen SK et al. RNA-sequencing of the brain transcriptome implicates dysregulation of neuroplasticity, circadian rhythms and GTPase binding in bipolar disorder. Mol Psychiatry 2014; 19: 1179–1185.
Weickert CS, Ligons DL, Romanczyk T, Ungaro G, Hyde TM, Herman MM et al. Reductions in neurotrophin receptor mRNAs in the prefrontal cortex of patients with schizophrenia. Mol Psychiatry 2005; 10: 637–650.
Hwang Y, Kim J, Shin JY, Kim JI, Seo JS, Webster MJ et al. Gene expression profiling by mRNA sequencing reveals increased expression of immune/inflammation-related genes in the hippocampus of individuals with schizophrenia. Transl Psychiatry 2013; 3: e321.
Zhao Z, Xu J, Chen J, Kim S, Reimers M, Bacanu SA et al. Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol Psychiatry 2015; 20: 563–572.
Bowden NA, Scott RJ, Tooney PA . Altered gene expression in the superior temporal gyrus in schizophrenia. BMC Genomics 2008; 9: 199.
Fatemi SH, Folsom TD, Rooney RJ, Thuras PD . Expression of GABAA alpha2-, beta1- and epsilon-receptors are altered significantly in the lateral cerebellum of subjects with schizophrenia, major depression and bipolar disorder. Transl Psychiatry 2013; 3: e303.
Hashimoto R, Straub RE, Weickert CS, Hyde TM, Kleinman JE, Weinberger DR . Expression analysis of neuregulin-1 in the dorsolateral prefrontal cortex in schizophrenia. Mol Psychiatry 2004; 9: 299–307.
Ryan MM, Lockstone HE, Huffaker SJ, Wayland MT, Webster MJ, Bahn S . Gene expression analysis of bipolar disorder reveals downregulation of the ubiquitin cycle and alterations in synaptic genes. Mol Psychiatry 2006; 11: 965–978.
Sinclair D, Webster MJ, Fullerton JM, Weickert CS . Glucocorticoid receptor mRNA and protein isoform alterations in the OFC in schizophrenia and bipolar disorder. BMC Psychiatry 2012; 12: 84.
Aberg KA, McClay JL, Nerella S, Clark S, Kumar G, Chen W et al. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry 2014; 71: 255–264.
Montano C, Taub MA, Jaffe A, Briem E, Feinberg JI, Trygvadottir R et al. Association of DNA methylation differences with schizophrenia in an epigenome-wide association study. JAMA Psychiatry 2016; 73: 506–514.
van den Oord EJ, Clark SL, Xie LY, Shabalin AA, Dozmorov MG, Kumar G et al. A whole methylome CpG-SNP association study of psychosis in blood and brain tissue. Schizophr Bull 2016; 42: 1018–1026.
Cai C, Langfelder P, Fuller TF, Oldham MC, Luo R, van den Berg LH et al. Is human blood a good surrogate for brain tissue in transcriptional studies? BMC Genomics 2010; 11: 589.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Torrey EF, Webster M, Knable M, Johnston N, Yolken RH . The Stanley foundation brain collection and neuropathology consortium. Schizophr Res 2000; 44: 151–155.
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL . TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14: R36.
Love MI, Huber W, Anders S . Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15: 550.
Merico D, Isserlin R, Stueker O, Emili A, Bader GD . Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 2010; 5: e13984.
Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S et al. A travel guide to Cytoscape plugins. Nat Methods 2012; 9: 1069–1076.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13: 2498–2504.
Mudge J, Miller NA, Khrebtukova I, Lindquist IE, May GD, Huntley JJ et al. Genomic convergence analysis of schizophrenia: mRNA sequencing reveals altered synaptic vesicular transport in post-mortem cerebellum. PLoS One 2008; 3: e3625.
Sawai H, Takai-Igarashi T, Tanaka H . Identification of collaborative activities with oxidative phosphorylation in bipolar disorder. Bioinformation 2015; 11: 207–216.
Focking M, Lopez LM, English JA, Dicker P, Wolff A, Brindley E et al. Proteomic and genomic evidence implicates the postsynaptic density in schizophrenia. Mol Psychiatry 2015; 20: 424–432.
Schubert KO, Focking M, Cotter DR . Proteomic pathway analysis of the hippocampus in schizophrenia and bipolar affective disorder implicates 14-3-3 signaling, aryl hydrocarbon receptor signaling, and glucose metabolism: potential roles in GABAergic interneuron pathology. Schizophr Res 2015; 167: 64–72.
Wikenheiser AM, Schoenbaum G . Over the river, through the woods: cognitive maps in the hippocampus and OFC. Nat Rev Neurosci 2016; 17: 513–523.
International Schizophrenia C, Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–752.
Topol A, English JA, Flaherty E, Rajarajan P, Hartley BJ, Gupta S et al. Increased abundance of translation machinery in stem cell-derived neural progenitor cells from four schizophrenia patients. Transl Psychiatry 2015; 5: e662.
Zubenko GS, Hughes HB 3rd, Jordan RM, Lyons-Weiler J, Cohen BM . Differential hippocampal gene expression and pathway analysis in an etiology-based mouse model of major depressive disorder. Am J Med Genet B Neuropsychiatr Genet 2014; 165B: 457–466.
Graber TE, Hebert-Seropian S, Khoutorsky A, David A, Yewdell JW, Lacaille JC et al. Reactivation of stalled polyribosomes in synaptic plasticity. Proc Natl Acad Sci USA 2013; 110: 16205–16210.
Heise C, Gardoni F, Culotta L, di Luca M, Verpelli C, Sala C . Elongation factor-2 phosphorylation in dendrites and the regulation of dendritic mRNA translation in neurons. Front Cell Neurosci 2014; 8: 35.
Duman RS, Li N, Liu RJ, Duric V, Aghajanian G . Signaling pathways underlying the rapid antidepressant actions of ketamine. Neuropharmacology 2012; 62: 35–41.
Horvath S, Mirnics K . Schizophrenia as a disorder of molecular pathways. Biol Psychiatry 2015; 77: 22–28.
Horvath S, Mirnics K . Immune system disturbances in schizophrenia. Biol Psychiatry 2014; 75: 316–323.
Khandaker GM, Cousins L, Deakin J, Lennox BR, Yolken R, Jones PB . Inflammation and immunity in schizophrenia: implications for pathophysiology and treatment. Lancet Psychiatry 2015; 2: 258–270.
Garbett KA, Vereczkei A, Kalman S, Brown JA, Taylor WD, Faludi G et al. Coordinated messenger RNA/microRNA changes in fibroblasts of patients with major depression. Biol Psychiatry 2015; 77: 256–265.
Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–752.
Morgan LZ, Rollins B, Sequeira A, Byerley W, DeLisi LE, Schatzberg AF et al. Quantitative trait locus and brain expression of HLA-DPA1 offers evidence of shared immune alterations in psychiatric disorders. Microarrays (Basel) 2016; 5: (doi:10.3390/microarrays5010006).
Choi KH, Elashoff M, Higgs BW, Song J, Kim S, Sabunciyan S et al. Putative psychosis genes in the prefrontal cortex: combined analysis of gene expression microarrays. BMC Psychiatry 2008; 8: 87.
Sequeira A, Morgan L, Walsh DM, Cartagena PM, Choudary P, Li J et al. Gene expression changes in the prefrontal cortex, anterior cingulate cortex and nucleus accumbens of mood disorders subjects that committed suicide. PLoS One 2012; 7: e35367.
Montalbetti N, Simonin A, Kovacs G, Hediger MA . Mammalian iron transporters: families SLC11 and SLC40. Mol Aspects Med 2013; 34: 270–287.
Xu J, Sun J, Chen J, Wang L, Li A, Helm M et al. RNA-Seq analysis implicates dysregulation of the immune system in schizophrenia. BMC Genomics 2011; 13 Suppl 8: S2.
Chen YH, Tsai MT, Shaw CK, Chen CH . Mutation analysis of the human NR4A2 gene, an essential gene for midbrain dopaminergic neurogenesis, in schizophrenic patients. Am J Med Genet 2001; 105: 753–757.
Xing G, Zhang L, Russell S, Post R . Reduction of dopamine-related transcription factors Nurr1 and NGFI-B in the prefrontal cortex in schizophrenia and bipolar disorders. Schizophr Res 2006; 84: 36–56.
Guillozet-Bongaarts AL, Hyde TM, Dalley RA, Hawrylycz MJ, Henry A, Hof PR et al. Altered gene expression in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol Psychiatry 2014; 19: 478–485.
Tatro ET, Everall IP, Masliah E, Hult BJ, Lucero G, Chana G et al. Differential expression of immunophilins FKBP51 and FKBP52 in the frontal cortex of HIV-infected patients with major depressive disorder. J Neuroimmune Pharmacol 2009; 4: 218–226.
Sinclair D, Fillman SG, Webster MJ, Weickert CS . Dysregulation of glucocorticoid receptor co-factors FKBP5, BAG1 and PTGES3 in prefrontal cortex in psychotic illness. Sci Rep 2013; 3: 3539.
Acknowledgements
We thank the Stanley Medical Research Institute for funding this research. We thank Drs E Fuller Torrey and Maree Webster for providing the samples. We also thank Ms Ou Chen for constructing libraries and performing the sequencing, and Mr Suad Diglisic and Ms Elizabeth Rubalcaba for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
RHY is a member of the Stanley Medical Research Institute Board of Directors and Scientific Advisory Board. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies. The remaining authors declare no conflicts of interest.
Additional information
Supplementary Information accompanies the paper on the Translational Psychiatry website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Darby, M., Yolken, R. & Sabunciyan, S. Consistently altered expression of gene sets in postmortem brains of individuals with major psychiatric disorders. Transl Psychiatry 6, e890 (2016). https://doi.org/10.1038/tp.2016.173
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2016.173
This article is cited by
-
What genes are differentially expressed in individuals with schizophrenia? A systematic review
Molecular Psychiatry (2022)
-
Amygdala and anterior cingulate transcriptomes from individuals with bipolar disorder reveal downregulated neuroimmune and synaptic pathways
Nature Neuroscience (2022)
-
Altered extracellular mRNA communication in postpartum depression is associated with decreased autophagy
Molecular Psychiatry (2022)
-
Differential expression of gene co-expression networks related to the mTOR signaling pathway in bipolar disorder
Translational Psychiatry (2022)
-
Increased levels of TAR DNA-binding protein 43 in the hippocampus of subjects with bipolar disorder: a postmortem study
Journal of Neural Transmission (2022)