Abstract
We present a 3D time-lapse imaging method for monitoring mitochondrial dynamics in living HeLa cells based on photothermal optical coherence microscopy and using novel surface functionalization of gold nanoparticles. The biocompatible protein-based biopolymer coating contains multiple functional groups which impart better cellular uptake and mitochondria targeting efficiency. The high stability of the gold nanoparticles allows continuous imaging over an extended time up to 3000 seconds without significant cell damage. By combining temporal autocorrelation analysis with a classical diffusion model, we quantify mitochondrial dynamics and cast these results into 3D maps showing the heterogeneity of diffusion parameters across the whole cell volume.
Similar content being viewed by others
Introduction
Mitochondria are organelles present in most eukaryotic cells. They are important for sustaining the energy needs of the host cell via the synthesis of ATP1. Besides being the cellular powerhouse, mitochondria are also involved in synaptic transmission and cellular signaling2. These fundamental functions translate into continuous mitochondria trafficking, which is an essential characteristic of these organelles, best subsumed by the term “mitochondrial dynamics”3. Quantifying these dynamics over a large time span promises to deepen our insight on cellular processes tightly related to neurodegenerative diseases such as Alzheimer’s and Parkinson’s disease1,4,5,6.
Confocal fluorescence microscopy remains the most widely used technique for cellular imaging. It provides high spatial resolution and specificity, known from fluorescent markers, making it suitable for intracellular imaging and studying mitochondria morphology. However, available fluorescent markers are prone to photobleaching, such as organic dyes, or are inherently toxic, like quantum dots7,8, limiting their viability for long-lasting live cell imaging.
Light-sheet micrsocopy is a fast, 3D, and high resolution intracellular imaging technique that also addresses photobleaching. In particular, Planchon, T. A. et al.9 demonstrated imaging live-cell mitochondrial dynamics using a scanned Bessel beam. Live pig kidney epithelial cells (LLC-PK1 cell line) were imaged over 300 volume stacks displaying a decrease in fluorescence intensity of only <20%. Among these techniques, probably the most advanced is lattice light-sheet microscopy by Chen, B.-C. et al.10 where imaging mitochondrial dynamics for more than 18 minutes was achieved with limited photobleaching. Undoubtedly, light-sheet microscopy has succeeded in pushing the bounds of intracellular imaging. However, it still relies on fluorescence probes and will eventually be confronted with the same limitations.
The use of gold nanoparticles (AuNPs) as biomarkers for photothermal imaging offers a promising alternative because of their high stability and very low toxicity. However, to accomplish photothermal imaging inside living cells, it is crucial to stabilize the surface of the AuNPs with water-soluble, biocompatible ligands that can withstand various physiological conditions. Stability of the AuNPs at varying pH and in the presence of proteases located in vesicles during endosomal uptake processes or in the reductive environment in the cytoplasm are essential to provide suitable intracellular markers for time lapse studies11. Protein based polymeric surface coatings offer the additional advantage as multiple functionalities could be incorporated at the level of the polymer. This ensures characterization by standard polymer analytics and guarantees the presence of all required functions at the particle surface after coating12. In this way, photothermal imaging AuNPs could be envisaged carrying the desired functionalities for targeting sub-cellular organelles, such as mitochondria, inside living cells.
The photothermal contrast mechanism relies on a temporally modulated refractive index in the near vicinity of the AuNPs resulting from heat dissipated by the AuNPs’ plasmon-enhanced absorption. This contrast mechanism is the basis for photothermal optical lock-in optical coherence microscopy (poli-OCM), which provides two distinct imaging modalities: a dark-field mode (dfOCM) for imaging the 3D cell volume, and a poli-mode, utilizing functionalized AuNPs for highly specific 3D mitochondria imaging13.
In this work, we report the synthesis of a novel and biocompatible protein-based biopolymer for surface functionalization of AuNPs. The biopolymer is comprised of multiple functional groups imparting enhanced cellular uptake and mitochondria targeting. Using these AuNPs, 3D mitochondria specific poli-OCM imaging during 3000 seconds was demonstrated without any loss of contrast. Finally, we quantified mitochondrial dynamics, per voxel, using temporal autocorrelation analysis based on a classical diffusion model, which allowed us to extract mitochondria diffusion time τD and other diffusion parameters. The novelty of our method resulted in cells segmented into sub-volumes providing 3D parameter maps.
Results and Discussion
Synthesis of mitochondria specific AuNPs
We synthesized mitochondria targeting AuNP biomarkers tailor-made for poli-OCM imaging. The synthesis, functionalization, and characterization of these AuNPs are summarized in Fig. 1a–d. The blood plasma protein is known to be biocompatible and provides many reactive carboxylic acid and amino groups that can be further modified. First, all accessible carboxylic acid groups were converted into primary amino groups by applying ethylenediamine and the coupling reagent EDC according to a literature-known procedure14. After dialysis, globular polycationic bovine serum albumin (cBSA) was obtained having the ability to interact with cellular membranes, facilitate cellular uptake, and cytosolic release by Clathrin-mediated endocytosis15. To accomplish mitochondria targeting, lipophilic triphenyl phosphonium groups (TPP) were attached to cBSA16. According to the MALDI-ToF mass spectra (Fig. 1b), about 19 TPP groups were attached to the cBSA surface yielding cBSA-TPP in good yields. cBSA-TPP was then purified and subjected to AuNP preparation. We used a strong reducing agent, NaBH4, to reduce Au-salt (HAuCl4) in the presence of cBSA-TPP and yielded monodispersed spherical AuNPs with diameters centered around 3.7 ± 0.9 nm (Fig. 1c). The inset histogram map confirms the relative narrow size distribution and Fig. 1d shows the characteristic plasmonic peak of the prepared AuNPs. Since the stability of the cBSA-TPP passivated AuNPs in biological media is crucial for cellular uptake and subsequent observation of their movement without prior aggregation, dynamic light scattering experiments were performed in DMEM medium (see Fig. S2b). No indication of severe aggregation, as little increase of the particle size, was observed, which is believed to be due to the alteration of ionic strength in biological medium as compared to its aqueous counterpart17.
(a) Synthesis scheme of globular cationized protein (cBSA-TPP) with multiple mitochondria targeting TPP groups attached. (b) MALDI-ToF spectra (matrix: sinapinic acid) indicate successful functionalization from the progressive increase in molecular weight from BSA (calculated 66.00 kDa, measured 66.13 kDa), cBSA (calculated 71.05 kDa, measured 71.04 kDa) and cBSA-TPP (calculated 77.62 kDa, measured 77.51 kDa). Approximately 19 TPP units were attached. (c) Low resolution transmission electron microscopy (TEM) image of cBSA-TPP coated AuNPs. Scalebar: 50 nm. Inset: Size distribution histogram indicating average diameters of the AuNPs of 3.7 ± 0.9 nm. (d) Characteristic absorption spectra of the as-synthesized AuNPs highlighting the surface plasmon peak centered at 536 nm.
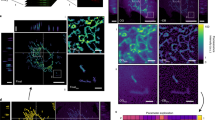
In order to demonstrate the specificity of our AuNP biomarkers, HeLa cells with AuNP labeled mitochondria were co-stained with MitoTracker Red, a standard mitochondria specific dye. These AuNPs were linked with FITC fluorophores which enabled imaging via confocal laser scanning microscopy and colocalization with MitoTracker staining. We conducted this test using two different kinds of AuNP functionalization; the first group of AuNP-cBSA was attached with TPP (AuNP-cBSA-TPP) while the second one was not (AuNP-cBSA). Figure 2a–f shows the fluorescence confocal images of the HeLa cells with MitoTracker (red), AuNP-FITC (green), and their overlays. From these images, we observed good colocalization of AuNP-cBSA-TPP with MitoTracker giving a Pearson’s coefficient of 0.69 (Fig. 2d–f) as opposed to 0.29 for AuNP-cBSA (Fig. 2a–c). These results are indicative of the specificity of our AuNP biomarkers and substantiate their use for mitochondria specific poli-OCM imaging. In addition, we also investigated the biocompatibility of our AuNP labels through a cell viability assay (Fig. 2g and Fig. S3). We obtain 95% cell viability at 0.31 nM AuNP concentration which is the typical AuNP labeling concentration we use for poli-OCM.
Fluorescence confocal images of HeLa cells incubated with AuNP-cBSA (a–c) and AuNP-cBSA-TPP (d–f) co-stained with MitoTracker. (a and d) are the MitoTracker images whereas (b and e) are the FITC tagged AuNPs. Their overlays in (c and f) have Pearson’s coefficients of 0.29 and 0.69 for AuNP-noTPP and AuNP-TPP respectively. Scalebars: 30 μm. (g) Cell viability test showing 95% viability HeLa cells incubated with 0.31 nM of mitochondria targeting AuNPs.
Quantifying mitochondrial dynamics
We quantified mitochondrial dynamics by exploiting fast and specific 3D imaging via poli-OCM and using a diffusion model considering the mitochondria as freely diffusing particles inside the voxel volume. Knowing the high complexity of the intracellular structure, we argue that using a voxel sized sampling volume allows the use of a diffusion model supposing a homogeneous environment. This model permits to assess the general mitochondrial dynamic and to cast our results in a parameter space known and used in correlation microscopy.
Our correlation analysis closely follows the principles of image correlation spectroscopy (ICS) developed by Petersen et al.18 and Wiseman et al.19,20 that extended fluorescence correlation spectroscopy (FCS) to full 2D imaging methods. ICS based correlation analysis on pixel-wise intensity fluctuations provide the diffusion time of the particles under investigation. Merging photothermal detection and correlation spectroscopy was previously introduced21,22 but not with 3D imaging or extended time scales (up to 1 hour)23. In a similar manner, phase correlation imaging has also been demonstrated as a robust technique for studying cell dynamics24 even without the use of specific labels. As we have previously stated, using the poli-OCM we achieved fast 3D mitochondria specific live cell imaging up to 3000 seconds without any loss of contrast (see Fig. S6 and Movie S1,S2,S3).
Stepping from 2D to 3D imaging, the voxel-wise autocorrelation function can be stated as

where the indices V point to the individual voxels i.e. the sampling volumes, 〈〉 denotes time averaging, iV(t) the time-dependent intensity and τ the temporal lag. The temporal intensity fluctuation is then defined as δiV(t) = iV(t) − 〈δiV(t + τ)〉. Assuming a 3D Gaussian sampling volume, (see Fig. S5), the processed intensity data are fitted with the model for classical 3D diffusion behavior

Here  where r0 and z0 represent the lateral and axial extent of the sampling volume, τD is the diffusion time, and G∞ = limτ → ∞GV(τ)20,25. In FCS,
where r0 and z0 represent the lateral and axial extent of the sampling volume, τD is the diffusion time, and G∞ = limτ → ∞GV(τ)20,25. In FCS,  , where N is the mean number of diffusing particles. G(0), in turn, is dependent on the signal to noise ratio and also considers “independent particles”26, which in the case of poli-OCM can be influenced by AuNP labelling efficiency and interparticle coupling27; an in depth analysis of this dependence is beyond the scope of this study. All instrument parameters, r0 and z0, were evaluated by measuring the point spread function (PSF), fitting it with a Gaussian profile, (Fig. S5) and by taking into account the coherence length which defines the axial extent of the PSF28,29. For a more in depth discussion of coherent correlation analysis please refer to previous papers in OCCS28,29. Due to the interferometric detection, our signal has extremely high sensitivity to phase and not only intensity in contrast to ICS of Wiseman and Petersen.
, where N is the mean number of diffusing particles. G(0), in turn, is dependent on the signal to noise ratio and also considers “independent particles”26, which in the case of poli-OCM can be influenced by AuNP labelling efficiency and interparticle coupling27; an in depth analysis of this dependence is beyond the scope of this study. All instrument parameters, r0 and z0, were evaluated by measuring the point spread function (PSF), fitting it with a Gaussian profile, (Fig. S5) and by taking into account the coherence length which defines the axial extent of the PSF28,29. For a more in depth discussion of coherent correlation analysis please refer to previous papers in OCCS28,29. Due to the interferometric detection, our signal has extremely high sensitivity to phase and not only intensity in contrast to ICS of Wiseman and Petersen.
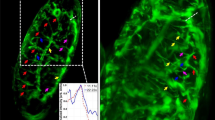
The dual imaging modality of the poli-OCM is exhibited in Fig. 3a–d where we show en face and orthogonal slices of the 3D dfOCM (Fig. 3a), photothermal OCM (Fig. 3b) tomograms, and their overlay (Fig. 3c). The orthogonal slices emphasize the 3D specific imaging and localization we achieve using the poli-OCM; we are capable of imaging the entire cell morphology (Fig. 3a) as well as only the AuNP labeled mitochondria (Fig. 3b). For the purposes of our temporal autocorrelation analysis we image individual cells in a scan range of 25 μm × 25 μm. Alternating between dfOCM and poli-mode, tomograms similar to (Fig. 3a and b) were acquired to monitor both the motion of the entire cell (dfOCM) and mitochondrial dynamics (photothermal OCM).
En face and orthogonal slices of (a) 3D dark-field OCM and (b) poli-OCM tomograms and (c) their overlay. In (d) we show selected autocorrelation data with corresponding fits and their residuals. Inset: normalized autocorrelation and fits. (e) Probability distribution of all extracted diffusion times from each sub-volumes in the entire cell is shown. The color-coded segments of (e) each represents approximately 20% of the complete cell volume. (f) 3D rendering (diffusion time map) using data from (e) illustrating the high heterogeneity of mitochondrial dynamics within the cell. The semi-transparent grey volume outlines the entire cell volume. (d–f) Shares the same color-code meaning each curve in (d) comes from the same colored segment in e) as well as the same colored sub-volume in (f) Scalebars: (a–c) 30 μm en face and 7.5 μm orthogonal slices, (f) 5 μm.
The temporal autocorrelation was calculated for each voxel in the 3D poli-OCM tomogram time series following equation (1). The data was then segmented into sub-volumes of 2 × 2 × 2 voxel (0.58 μm × 0.58 μm × 2.05 μm) where the autocorrelation was averaged and subsequently fitted with the diffusion model given by equation (2) using a non-linear least squares solver. Our method allowed the extraction of diffusion parameters from each of these segmented sub-volumes resulting in a 3D diffusion time map (Fig. 3f). Furthermore, the 2 × 2 × 2 voxels sub-volume we use are related to the measured dimensions of our point spread function (PSF).
The data shown in Fig. 3d represent five sub-volumes with different diffusion times (see matched color code of histogram in Fig. 3e) underlining the high variation of the mitochondrial dynamics across the full cell volume. The very small residuals between the autocorrelation and the model indicate a good fit and give confidence to the validity of the model. Additionally, the normalized autocorrelations (Fig. 3d, inset) demonstrate even more the large variation of τD. The probability distribution (pdf) of the τD extracted from all sub-volumes in the entire cell (Fig. 3e) demonstrates a spread of over two orders of magnitude with a τD ranging from 10 to 1.5 × 103 seconds having a mean value of 214 sec. (median value of 101 sec.). We divided the pdf (τD) into five color-coded intervals, each representing 20% of the total cell volume (Fig. 3e). These intervals were used to render a 3D map of the diffusion time, as shown in Fig. 3f and Fig. S9 with each sub-volume color-coded according to the pdf (τD). This 3D map allows us to determine and locate regions within the cell featuring high or low mitochondrial dynamics. These extracted τD can be further related, by the Stokes-Einstein relation (see Fig. S8), to the viscosity values inside our sampling volumes. Assuming a mitochondria hydrodynamic radius of 500 nm we deduced viscosity values ranging from 0.18 to 26.7 (Pa⋅s), which are still within the range predicted by the model of Kalwarczyk et al.30. In addition, the associated diffusion constants we calculate from these data coincide with published quantified cellular dynamics using phase correlation imaging24.
In addition to the high heterogeneity of mitochondrial dynamics, regions within the cell, where it was not possible to extract any diffusion parameters were also observed. As elaborated in Section 6 of the Supporting Information, this region exhibits a high overlap with the locations in the corresponding dfOCM (dark-field OCM) time series showing lower scattering signal. We associate this low scattering regions to the nucleus of the cell where no mitochondria are expected.
The high heterogeneity of diffusion parameters over the cell volume is an observed phenomenon resulting from our data and analysis. This is not surprising given the known complexity of the intracellular environment31,32,33, especially considering the size of mitochondria and their tendency to form networks. Furthermore, the works of Planchon et al.9 and Chen, B.-C. et al.10 also observed this variety in mitochondrial dynamics using a completely different and independent imaging concept. We would like to reiterate that, unlike fluorescence, poli-OCM imaging is not prone to photobleaching at all; as shown in Fig. S6, there is no decrease in signal contrast even after acquiring 500 poli-OCM tomograms. We, nevertheless, consider lattice light-sheet microscopy as the golden standard for imaging live intracellular dynamics and poli-OCM imaging compares well with these published time-lapse acquisition protocols. These previous works in light sheet microscopy9,10 demonstrated similar high heterogeneity of mitochondrial dynamics and can therefore be considered as an independent validation of our observations.
Conclusion
In this paper, we report highly specific fast 3D imaging of mitochondrial dynamics inside living HeLa cells. We prepared narrowly dispersed AuNPs with a protein coating based on the plasma protein BSA, functionalized with positive ammonium and multiple TPP groups. The positive ammonium groups improve cell membrane attachment and endocytosis while the TPP groups provide intracellular mitochondria targeting. All AuNPs used in this study were decorated by the functionalized protein coating approach. The photostability and limited toxicity of these markers enabled true long-lasting time lapse live cell imaging. Finally, we used these functionalized AuNPs for fast 3D poli-OCM imaging to quantify mitochondrial dynamics via temporal autocorrelation analysis resulting to 3D diffusion parameters maps. We believe that this method provides a novel perspective of mitochondrial dynamics and paves an innovative way for investigating its relation to various not yet understood metabolic diseases.
Methods
Preparation of functionalized protein coated AuNPs
Materials used
Cationized bovine serum albumin (cBSA) was synthesized following the procedure reported previously by our group34 N-hydroxy-succinimide (NHS, 99%), (3-Carboxypropyl) triphenylphosphonium bromide (TPP), 98% and N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC, 98%) were procured from Sigma-Aldrich. Hydrogen tetrachloro aurate (III) trihydrate, ACS, 99.99% was obtained from Alfa Aesar. Sodium borohydride 98% was bought from Acros Organics. All chemicals were used as received without further purification. Vivaspin ultrafiltration tubes were purchased from GE healthcare. Ultra-pure milli-Q water was used for all experiments involving water.
Synthesis of cBSA-TPP
TPP (80 mg), NHS (30 mg) and EDC.HCl (40 mg) were dissolved in 0.5 mL of degassed dimethylformamide (DMF) solution. This mixture was stirred at room temperature under argon atmosphere for overnight. Next day, cBSA (20 mg) dissolved in 20 mL milli-Q water was added and reacted overnight at room temperature. The product was washed through vivaspin 20 (MWCO 30 K) ultracentrifuge tube to separate the unreacted reactants. Finally, the solution was kept at 4 °C for future use. Figure S1 shows the zeta potential and XPS data of the conjugate.
Synthesis of cBSA-TPP coated AuNPs
AuNPs with desired sizes were synthesized and functionalized for mitochondria labeling. Briefly, aqueous HAuCl4 solution (60 μL, 10 mM) and cBSA-TPP (globular cationic bovine serum albumin with triphenyl phosphonium cations, 50 μL, 10 mg/mL) were mixed and the solution was diluted to 250 μL with milli-Q water. Freshly prepared NaBH4 solution (6 μL, 3 mg/mL) in chilled milli-Q water was added rapidly to the solution which yielded monodispersed, spherical AuNPs with diameter of approximately 4 nm. Then, the reaction mixture was stirred for 30 minutes and washed with water to remove excess of NaBH4. By varying the amount of added NaBH4, various sizes of the AuNPs were obtained. In particular, the AuNPs used in the poli-OCM imaging experiments had diameters of 3.7 ± 0.9 nm. Corresponding XPS peaks of AuNPs are presented in Fig. S2a.
AuNP characterization
Plasmonic absorption was recorded using TECAN infinite M1000 microplate reader. Zeta-potential and DLS measurements were performed using a Malvern Zetasizer ZEN3600 (Malvern Ltd, Malvern, UK) at 20. A JEOL 1400 transmission electron microscope was used to obtain bright field TEM images of the AuNPs. X-ray photoelectron spectroscopy (XPS) data were recorded on a Physical Electronics PHI 5800 ESCA System using mono-chromatized Al Kα radiation (13 kV, 250 W).
Cell viability assay
HeLa cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) with 10% fetal bovine serum, 1% penicillin/streptomycin with phenol-red and seeded at 6,500 cells/well in a white 96-well (half-area) plate. The cells were left to adhere overnight at 37 °C and 5% CO2 and afterwards different concentrations of AuNPs were added into each well. The treated cells were subsequently incubated with the AuNPs for approximately 30 hours at 37 °C, 5% CO2. After incubation, the cells were washed with phosphate buffer to remove the excess AuNPs present in the medium and re-incubated with DMEM medium. The cells were further incubated for 4 hours before the addition of Tox-8 reagent with phenol red free DMEM medium. After 2 hours incubation with Tox-8, the emission intensity was measured by a Tecan Infinite M1000 microplate reader (λex = 570 nm, λem = 590 nm). We used wells without cells but with Tox-8 reagent as controls. Each experiment was performed in quadruplicates. The cell viability (V) was calculated according to the following equation

where X is the average emission with the treatment of AuNPs at various concentrations, Y is the average emission of the experimental groups without the treatment of the AuNPs, and Z is the average emission of the culture medium background.
Confocal microscopy
HeLa cells were incubated in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) with 10% fetal calf serum (FCS, Gibco) and non-essential amino acids in a humidified incubator at 37 °C and 5% CO2. For confocal laser scanning microscopy (CLSM), cells were seeded onto an Ibidi μ-slide 8-well and incubated with FITC labelled protein coated AuNPs for 24 hours. We used two different kinds of protein coating: one with TPP attached and the other without TPP attached in its backbone. Fluorescence confocal laser scanning microscopy (CLSM) images were acquired with a 63 × 1.4 NA oil immersion plan apochromatic LSM 710, Axio Observer objective. Fluorescence of FITC was excited by a HeNe laser λex = 488 nm (AOTF transmission set to 10–20%) and was set to emission maximum at 525 nm. We added 1 μM MitoTracker Red in each well-plate and excited at 560 nm with an emission maximum at 625 nm.
poli-OCM imaging
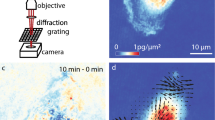
The 3D time-lapse imaging and analysis we implemented for the quantitative assessment of mitochondrial dynamics is summarized schematically in Fig. 4.
3D time lapse imaging of live HeLa cells was performed using the poli-OCM localizing the AuNP labeled mitochondria. Temporal autocorrelation analysis was applied over the whole cell volume resulting in a 3D map of mitochondria diffusion time. In addition, imaging with the poli-OCM also enables mitochondrial dynamics quantification via tracking individual AuNP-labeled mitochondria but is outside the scope of this work.
Cell culture and AuNP labelling
HeLa cells were seeded and incubated overnight onto an Ibidi μ-slide 8-well with 250 μL of cell culture medium (Dulbecco’s Modified Eagle Medium (DMEM, Gibco), 10% fetal bovine serum, and 1% penicillin and streptomycin antibiotics). 2 μL of 40 nM AuNP solution were then added into the media and again incubated overnight. Finally, the cells were washed with PBS and fresh culture medium was added before imaging. During poli-OCM imaging, the Ibidi μ-slide 8-well was housed in a custom built micro-incubator, where the temperature was regulated at 37 °C and humidified premixed (5% CO2) air was pumped continuously.
poli-OCM Instrumentation
The specifics of the OCM instrument as shown in Fig. S4 have been previously described and characterized elsewhere (xfOCM35, dfOCM36 and poli-OCM13). A broadband laser source centered at 800 nm (Δλ = 135 nm) is used as a source for optical coherence imaging. An axicon shapes the illumination beam into a Bessel beam which propagates across the scanning unit and is focused on the sample by a high-NA objective (25×, NA = 0.8, Carl Zeiss). The backscattered field is collected by the same objective and superimposed with the reference beam; the resulting interference is finally recorded using a custom-made spectrometer. The Bessel beam configuration of our poli-OCM ensures an extended depth of field or a uniform lateral resolution over the full 3D cellular volume. The poli-OCM, as used in this study, provides a lateral and axial resolution of 0.53 μm and 2.15 μm respectively over an extended depth of focus of 50 μm. The experimental measurement of the resolution is discussed in the supporting information. The photothermal contrast of the poli-OCM13 is achieved by exciting the plasmonic resonance of the AuNPs using a 532 nm solid-state laser. This photothermal heating beam is intensity modulated at 150 kHz using an AOM before being coupled into the OCM scanning unit by a dichroic mirror (D1; 720DCXR, Chroma Technology). This intensity modulation induces a similarly modulated index of refraction around the AuNP which in turn generates a modulated backscattered signal. This backscattered field is superimposed with a phase modulated reference field (150 kHz) enabling heterodyne detection of the sinusoidally modulated signal. In accordance with C. Pache et al. we operate at an A-scan rate of 3900 Hz with an integration time of 250 μs13. At this operating parameters the temperature increase in the vicinity of an individual AuNP is approximately 1 K13,37. During imaging, we used approximately 6 mW for OCM probe beam and 3 mW average power for the photothermal heating laser.
Time lapse imaging
Aiming for the extraction of diffusion parameters of the AuNP labeled mitochondria requires fast 3D imaging with short time intervals while extending the experiment over a prolonged acquisition period (100 to 103 seconds). A total of 1000 3D tomograms were acquired over a volume of 25 × 25 × 50 μm3 (approximately 0.29 × 0.29 × 1.028 μm3 per voxel) while alternating between dfOCM and poli-OCM. This resulted in two time series each having 500 3D tomograms and a time interval of 6 seconds between tomograms. This scan protocol provided us with sufficient sampling necessary for our autocorrelation analysis. Similarly, poli-OCM imaging allows for 3D tracking of AuNP labeled mitochondria but this is not within the scope of this current work.
Additional Information
How to cite this article: Sison, M. et al. 3D Time-lapse Imaging and Quantification of Mitochondria Dynamics. Sci. Rep. 7, 43275; doi: 10.1038/srep43275 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Burte, F., Carelli, V., Chinnery, P. F. & Yu-Wai-Man, P. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nature Reviews Neurology 11, 11–24 (2015).
Sheng, Z. H. & Cai, Q. Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nature Reviews Neuroscience 13, 77–93 (2012).
Detmer, S. A. & Chan, D. C. Functions and dysfunctions of mitochondrial dynamics. Nature Reviews Molecular Cell Biology 8, 870–879 (2007).
Chan, D. C. Mitochondria: Dynamic organelles in disease, aging, and development. Cell 125, 1241–1252 (2006).
Barnham, K. J., Masters, C. L. & Bush, A. I. Neurodegenerative diseases and oxidative stress. Nature reviews. Drug discovery 3, 205–14 (2004).
Lin, M. T. & Beal, M. F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443, 787–95 (2006).
Chakraborty, A. & Jana, N. R. Design and Synthesis of Triphenylphosphonium Functionalized Nanoparticle Probe for Mitochondria Targeting and Imaging. The Journal of Physical Chemistry C150127145409002 (2015).
Fanizza, E. et al. Highly selective luminescent nanostructures for mitochondrial imaging and targeting. Nanoscale 8, 3350–3361 (2016).
Planchon, T. A. et al. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nature Methods 8, 417–423 (2011).
Chen, B.-C. et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science 346 (2014).
Boisselier, E. & Astruc, D. Gold nanoparticles in nanomedicine: preparations, imaging, diagnostics, therapies and toxicity. Chemical Society Reviews 38, 1759–1782 (2009).
Wu, Y. Z. et al. Programmable Biopolymers for Advancing Biomedical Applications of Fluorescent Nanodiamonds. Advanced Functional Materials 25, 6576–6585 (2015).
Pache, C. et al. Fast three-dimensional imaging of gold nanoparticles in living cells with photothermal optical lock-in Optical Coherence Microscopy. Optics express 20, 21385–21399 (2012).
Wu, Y. et al. A core-shell albumin copolymer nanotransporter for high capacity loading and two-step release of doxorubicin with enhanced anti-leukemia activity. Advanced healthcare materials 2, 884–94 (2013).
Eisele, K. et al. Fine-tuning DNA/albumin polyelectrolyte interactions to produce the efficient transfection agent cBSA-147. Biomaterials 31, 8789–8801 (2010).
Vayalil, P. K. et al. A Novel Class of Mitochondria-Targeted Soft Electrophiles Modifies Mitochondrial Proteins and Inhibits Mitochondrial Metabolism in Breast Cancer Cells through Redox Mechanisms. Plos One 10 (2015).
Sabuncu, A. C. et al. Probing nanoparticle interactions in cell culture media. Colloids and Surfaces B: Biointerfaces 95, 96–102 (2012).
Petersen, N., Höddelius, P., Wiseman, P., Seger, O. & Magnusson, K. Quantitation of membrane receptor distributions by image correlation spectroscopy: concept and application. Biophysical Journal 65, 1135–1146 (1993).
Wiseman, P. W., Squier, J. A., Ellisman, M. H. & Wilson, K. R. Two-photon image correlation spectroscopy and image cross-correlation spectroscopy. Journal of Microscopy 200, 14–25 (2000).
Wiseman, P. W. Spatial mapping of integrin interactions and dynamics during cell migration by Image Correlation Microscopy. Journal of Cell Science 117, 5521–5534 (2004).
Paulo, P. M. R. et al. Photothermal Correlation Spectroscopy of Gold Nanoparticles in Solution. Journal of Physical Chemistry C 113, 11451–11457 (2009).
Octeau, V. et al. Photothermal Absorption Correlation Spectroscopy. Acs Nano 3, 345–350 (2009).
Nieves, D. J., Li, Y., Fernig, D. G. & Levy, R. Photothermal raster image correlation spectroscopy of gold nanoparticles in solution and on live cells. Royal Society Open Science 2 (2015).
Ma, L. H. et al. Phase correlation imaging of unlabeled cell dynamics. Scientific Reports 6 (2016).
Wohland, T., Rigler, R. & Vogel, H. The standard deviation in fluorescence correlation spectroscopy. Biophysical Journal 80, 2987–2999 (2001).
Kolin, D. L. & Wiseman, P. W. Advances in image correlation spectroscopy: Measuring number densities, aggregation states, and dynamics of fluorescently labeled macromolecules in cells. Cell Biochemistry and Biophysics 49, 141–164 (2007).
Su, K. H. et al. Interparticle coupling effects on plasmon resonances of nanogold particles. Nano Letters 3, 1087–1090 (2003).
Broillet, S. et al. Optical coherence correlation spectroscopy (OCCS). Optics express 22, 782–802 (2014).
Broillet, S. et al. Visible light optical coherence correlation spectroscopy. Optics Express 22, 21944 (2014).
Kalwarczyk, T. et al. Comparative Analysis of Viscosity of Complex Liquids and Cytoplasm of Mammalian Cells at the Nanoscale (2011).
Luby-Phelps, K. Cytoarchitecture and Physical Properties of Cytoplasm: Volume, Viscosity, Diffusion, Intracellular Surface Area. In International Review of Cytology 192, 189–221 (1999).
Ellis, R. Macromolecular crowding: obvious but underappreciated. Trends in Biochemical Sciences 26, 597–604 (2001).
Ellis, R. Macromolecular crowding: an important but neglected aspect of the intracellular environment. Current Opinion in Structural Biology 11, 114–119 (2001).
Wu, Y., Pramanik, G., Eisele, K. & Weil, T. Convenient approach to polypeptide copolymers derived from native proteins. Biomacromolecules 13, 1890–8 (2012).
Leitgeb, R. A., Villiger, M., Bachmann, A. H., Steinmann, L. & Lasser, T. Extended focus depth for Fourier domain optical coherence microscopy. Optics Letters 31, 2450 (2006).
Leitgeb, R., Hitzenberger, C. & Fercher, A. Performance of fourier domain vs time domain optical coherence tomography. Optics Express 11, 889 (2003).
Berciaud, S., Lasne, D., Blab, G. A., Cognet, L. & Lounis, B. Photothermal heterodyne imaging of individual metallic nanoparticles: Theory versus experiment. Physical Review B 73, 045424 (2006).
Acknowledgements
The authors are grateful to T. Wohland for helpful hints and comments for this paper. This work has been partially supported by the ERC Synergy Grant 319130-BioQ grant (TW), the Swiss National Science Foundation and the German Research Foundation (DFG) under the DACH program (grants 205321L_135353 & 205320L_150191, TL & TW).
Author information
Authors and Affiliations
Contributions
M.S. and S.C. conducted the experiments and contributed equally to this work. M.S., S.C., and A.N. analysed the results. M.S., P.J.M. and J.E. reconfigured the poli-OCM. A.L. and M.S. built the customized incubator for live cell imaging. M.S., S.C., J.E., A.N., T.W. and T.L. conceived the experiments. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Sison, M., Chakrabortty, S., Extermann, J. et al. 3D Time-lapse Imaging and Quantification of Mitochondrial Dynamics. Sci Rep 7, 43275 (2017). https://doi.org/10.1038/srep43275
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep43275
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.