Abstract
Dendritic spines, the postsynaptic compartments of excitatory neurotransmission, have different shapes classified from ‘stubby’ to ‘mushroom-like’. Whereas mushroom spines are essential for adult brain function, stubby spines disappear during brain maturation. It is still unclear whether and how they differ in protein composition. To address this, we combined electron microscopy and quantitative biochemistry with super-resolution microscopy to annotate more than 47,000 spines for more than 100 synaptic targets. Surprisingly, mushroom and stubby spines have similar average protein copy numbers and topologies. However, an analysis of the correlation of each protein to the postsynaptic density mass, used as a marker of synaptic strength, showed substantially more significant results for the mushroom spines. Secretion and trafficking proteins correlated particularly poorly to the strength of stubby spines. This suggests that stubby spines are less likely to adequately respond to dynamic changes in synaptic transmission than mushroom spines, which possibly explains their loss during brain maturation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD015308. Image data are available from the corresponding author upon reasonable request.
Code availability
Code is available on GitHub: https://github.com/MSHelm/Nanomap.
References
Humeau, Y. & Choquet, D. The next generation of approaches to investigate the link between synaptic plasticity and learning. Nat. Neurosci. 22, 1536–1543 (2019).
Bourne, J. N. & Harris, K. M. Balancing structure and function at hippocampal dendritic spines. Annu. Rev. Neurosci. 31, 47–67 (2008).
Forrest, M. P., Parnell, E. & Penzes, P. Dendritic structural plasticity and neuropsychiatric disease. Nat. Rev. Neurosci. 19, 215–234 (2018).
Berry, K. P. & Nedivi, E. Spine dynamics: are they all the same? Neuron 96, 43–55 (2017).
Harris, K. M., Jensen, F. E. & Tsao, B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. J. Neurosci. 12, 2685–2705 (1992).
Spacek, J. & Harris, K. M. Three-dimensional organization of smooth endoplasmic reticulum in hippocampal CA1 dendrites and dendritic spines of the immature and mature rat. J. Neurosci. 17, 190–203 (1997).
Tønnesen, J., Katona, G., Rózsa, B. & Nägerl, U. V. Spine neck plasticity regulates compartmentalization of synapses. Nat. Neurosci. 17, 678–685 (2014).
Araya, R. Input transformation by dendritic spines of pyramidal neurons. Front. Neuroanat. https://doi.org/10.3389/fnana.2014.00141 (2014).
Nakahata, Y. & Yasuda, R. Plasticity of spine structure: local signaling, translation and cytoskeletal reorganization. Front. Synaptic Neurosci. 10, 29 (2018).
Biederer, T., Kaeser, P. S. & Blanpied, T. A. Transcellular nanoalignment of synaptic function. Neuron 96, 680–696 (2017).
Patriarchi, T., Buonarati, O. R. & Hell, J. W. Postsynaptic localization and regulation of AMPA receptors and Cav1.2 by β2 adrenergic receptor/PKA and Ca2+/CaMKII signaling. EMBO J. 37, e99771 (2018).
Papa, M., Bundman, M. C., Greenberger, V. & Segal, M. Morphological analysis of dendritic spine development in primary cultures of hippocampal neurons. J. Neurosci. 15, 1–11 (1995).
Kaech, S. & Banker, G. Culturing hippocampal neurons. Nat. Protoc. 1, 2406–2415 (2006).
Richter, K. N. et al. Glyoxal as an alternative fixative to formaldehyde in immunostaining and super‐resolution microscopy. EMBO J. https://doi.org/10.15252/embj.201695709 (2017).
Hruska, M., Henderson, N., Le Marchand, S. J., Jafri, H. & Dalva, M. B. Synaptic nanomodules underlie the organization and plasticity of spine synapses. Nat. Neurosci. https://doi.org/10.1038/s41593-018-0138-9 (2018).
Ikeda, K. & Bekkers, J. M. Counting the number of releasable synaptic vesicles in a presynaptic terminal. Proc. Natl Acad. Sci. USA 106, 2945–2950 (2009).
Schanzenbächer, C. T., Sambandan, S., Langer, J. D. & Schuman, E. M. Nascent proteome remodeling following homeostatic scaling at hippocampal synapses. Neuron 92, 358–371 (2016).
Schanzenbächer, C. T., Langer, J. D. & Schuman, E. M. Time- and polarity-dependent proteomic changes associated with homeostatic scaling at central synapses. eLife 7, e33322 (2018).
Ziv, N. E. & Brenner, N. Synaptic tenacity or lack thereof: spontaneous remodeling of synapses. Trends Neurosci. 41, 89–99 (2018).
Urban, N. T., Willig, K. I., Hell, S. W. & Nägerl, U. V. STED nanoscopy of actin dynamics in synapses deep inside living brain slices. Biophys. J. 101, 1277–1284 (2011).
Nägerl, U. V., Willig, K. I., Hein, B., Hell, S. W. & Bonhoeffer, T. Live-cell imaging of dendritic spines by STED microscopy. Proc. Natl Acad. Sci. USA 105, 18982–18987 (2008).
Willig, K. I. et al. Nanoscopy of filamentous actin in cortical dendrites of a living mouse. Biophys. J. 106, L01–L03 (2014).
Wegner, W. et al. In vivo mouse and live cell STED microscopy of neuronal actin plasticity using far-red emitting fluorescent proteins. Sci. Rep. 7, 11781 (2017).
Wegner, W., Mott, A. C., Grant, S. G. N., Steffens, H. & Willig, K. I. In vivo STED microscopy visualizes PSD95 sub-structures and morphological changes over several hours in the mouse visual cortex. Sci. Rep. 8, 219 (2018).
Turrigiano, G. G., Leslie, K. R., Desai, N. S., Rutherford, L. C. & Nelson, S. B. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature 391, 892–896 (1998).
Gallimore, A. R., Kim, T., Tanaka-Yamamoto, K. & De Schutter, E. Switching on depression and potentiation in the cerebellum. Cell Rep. 22, 722–733 (2018).
Freer, R. et al. Supersaturated proteins are enriched at synapses and underlie cell and tissue vulnerability in Alzheimer’s disease. Heliyon https://doi.org/10.1016/j.heliyon.2019.e02589 (2019).
Wilhelm, B. G. et al. Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science 344, 1023–1028 (2014).
Kirchhausen, T., Owen, D. & Harrison, S. C. Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb. Perspect. Biol. 6, a016725 (2014).
McMahon, H. T. & Boucrot, E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12, 517–533 (2011).
Pearse, B. M. & Crowther, R. A. Structure and assembly of coated vesicles. Annu. Rev. Biophys. Biophys. Chem. 16, 49–68 (1987).
Blanpied, T. A., Scott, D. B. & Ehlers, M. D. Dynamics and regulation of clathrin coats at specialized endocytic zones of dendrites and spines. Neuron 36, 435–449 (2002).
Esteves da Silva, M. et al. Positioning of AMPA receptor-containing endosomes regulates synapse architecture. Cell Rep. 13, 933–943 (2015).
Shnyrova, A. V. et al. Geometric catalysis of membrane fission driven by flexible dynamin rings. Science 339, 1433–1436 (2013).
Zheng, K. et al. Time-resolved imaging reveals heterogeneous landscapes of nanomolar Ca2+ in neurons and astroglia. Neuron 88, 277–288 (2015).
Higley, M. J. & Sabatini, B. L. Calcium signaling in dendritic spines. Cold Spring Harb. Perspect. Biol. 4, a005686 (2012).
Park, B. J. et al. Calreticulin, a calcium-binding molecular chaperone, is required for stress response and fertility in Caenorhabditis elegans. Mol. Biol. Cell 12, 2835–2845 (2001).
Nakamura, K. et al. Functional specialization of calreticulin domains. J. Cell Biol. 154, 961–972 (2001).
Solovyova, N., Veselovsky, N., Toescu, E. C. & Verkhratsky, A. Ca2+ dynamics in the lumen of the endoplasmic reticulum in sensory neurons: direct visualization of Ca2+-induced Ca2+ release triggered by physiological Ca2+ entry. EMBO J. 21, 622–630 (2002).
Biever, A., Donlin-Asp, P. G. & Schuman, E. M. Local translation in neuronal processes. Curr. Opin. Neurobiol. 57, 141–148 (2019).
Dörrbaum, A. R., Kochen, L., Langer, J. D. & Schuman, E. M. Local and global influences on protein turnover in neurons and glia. eLife 7, e34202 (2018).
Riba, A. et al. Protein synthesis rates and ribosome occupancies reveal determinants of translation elongation rates. Proc. Natl Acad. Sci. USA 116, 15023–15032 (2019).
Alberghina, F. A. M., Sturani, E. & Gohlke, J. R. Levels and rates of synthesis of ribosomal ribonucleic acid, transfer ribonucleic acid, and protein in Neurospora crassa in different steady states of growth. J. Biol. Chem. 250, 4381–4388 (1975).
Forchhammer, J. & Lindahl, L. Growth rate of polypeptide chains as a function of the cell growth rate in a mutant of Escherichia coli 15. J. Mol. Biol. 55, 563–568 (1971).
Miller, S. et al. Disruption of dendritic translation of CaMKIIα impairs stabilization of synaptic plasticity and memory consolidation. Neuron 36, 507–519 (2002).
Richter, K. N. et al. Comparative synaptosome imaging: a semi-quantitative method to obtain copy numbers for synaptic and neuronal proteins. Sci. Rep. 8, 14838 (2018).
Zhu, F. et al. Architecture of the mouse brain synaptome. Neuron 99, 781–799.e10 (2018).
Ciryam, P., Tartaglia, G. G., Morimoto, R. I., Dobson, C. M. & Vendruscolo, M. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep. 5, 781–790 (2013).
Fornasiero, E. F. et al. Precisely measured protein lifetimes in the mouse brain reveal differences across tissues and subcellular fractions. Nat. Commun. 9, 4230 (2018).
Mandad, S. et al. The codon sequences predict protein lifetimes and other parameters of the protein life cycle in the mouse brain. Sci. Rep. 8, 16913 (2018).
Turrigiano, G. G. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell 135, 422–435 (2008).
Matsuda, T. & Cepko, C. L. Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl Acad. Sci. USA 104, 1027–1032 (2007).
Schikorski, T. & Stevens, C. F. Quantitative ultrastructural analysis of hippocampal excitatory synapses. J. Neurosci. 17, 5858–5867 (1997).
Collins, M. O. et al. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J. Neurochem. 97, 16–23 (2006).
De Chaumont, F. et al. Icy: an open bioimage informatics platform for extended reproducible research. Nat. Methods 9, 690–696 (2012).
Olivo-Marin, J. C. Extraction of spots in biological images using multiscale products. Pattern Recognit. 35, 1989–1996 (2002).
Wishart, D. M. G. in Numerical Taxonomy (ed. Cole, A. J.) 282–311 (Academic Press, 1969).
Witkin, A. Scale-space filtering: a new approach to multi-scale description. In Proc. ICASSP ’84. IEEE International Conference on Acoustics, Speech, and Signal Processing 9 150–153 (Institute of Electrical and Electronics Engineers, 1983).
Leung, Yee, Zhang, Jiang-She & Xu, Zong-Ben Clustering by scale-space filtering. IEEE Trans. Pattern Anal. Mach. Intell. 22, 1396–1410 (2000).
Leibrandt, R. & Günnemann, S. Gauss Shift: Density Attractor Clustering Faster than Mean Shift. In Proc. ECML-PKDD 2020: The European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Databases (ECML-PKDD) 14–18 (2020).
Fukunaga, K. & Hostetler, L. The estimation of the gradient of a density function, with applications in pattern recognition. IEEE Trans. Inf. Theory 21, 32–40 (1975).
Smith, P. K. et al. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 (1985).
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Jahn, R., Schiebler, W., Ouimet, C. & Greengard, P. A 38,000-dalton membrane protein (p38) present in synaptic vesicles. Proc. Natl Acad. Sci. USA 82, 4137–4141 (1985).
Bohil, A. B., Robertson, B. W. & Cheney, R. E. Myosin-X is a molecular motor that functions in filopodia formation. Proc. Natl Acad. Sci. USA 103, 12411–12416 (2006).
Cohen, L. S. & Studzinski, G. P. Correlation between cell enlargement and nucleic acid and protein content of hela cells in unbalanced growth produced by inhibitors of DNA synthesis. J. Cell. Physiol. 69, 331–339 (1967).
Krombach, F. et al. Cell size of alveolar macrophages: an interspecies comparison. Environ. Health Perspect. 105, 1261–1263 (1997).
Luby-Phelps, K. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int. Rev. Cytol. 192, 189–221 (2000).
Puck, T. T., Marcus, P. I. & Cieciura, S. J. Clonal growth of mammalian cells in vitro; growth characteristics of colonies from single HeLa cells with and without a feeder layer. J. Exp. Med. 103, 273–283 (1956).
Zhao, L. et al. Intracellular water-specific MR of microbead-adherent cells: the HeLa cell intracellular water exchange lifetime. NMR Biomed. 21, 159–164 (2008).
Philips, R., Kodev, J., Therio, J. & Garcia, H. Physical Biology of the Cell (Garland Science, 2008).
Zou, M., Holloway, M., Carr, N. & Ju, T. Topology-constrained surface reconstruction from cross-sections. ACM Trans. Graph. https://doi.org/10.1145/2766976 (2015).
Roy, A., Kucukural, A. & Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. https://doi.org/10.1038/nprot.2010.5 (2010).
Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics https://doi.org/10.1186/1471-2105-9-40 (2008).
Acknowledgements
We thank J. Lovric and M. von Elling-Tamen for help with the immunostaining experiments, as well as N. Hartelt and C. Schäfer for expert technical assistance. We thank N. Brose (Göttingen, Germany), V. Haucke (Berlin, Germany), R. Jahn (Göttingen, Germany), P. Jonas (Klosterneuburg, Austria) and V. Nägerl (Bordeaux, France) for advice on the initial manuscript. We would also like to thank E. Fornasiero for many fruitful discussions and comments on the manuscript. The work was supported by a grant to S.O.R. from the Deutsche Forschungsgemeinschaft (DFG; SFB1286/A3, RI 1967/11-1, RI 1967/10-1/NeuroNex; SFB1190/P09, and under Germany’s Excellence Strategy - EXC 2067/1-390729940), by a grant to S.O.R. from Nieders Vorab (76251-12-6/19/ZN 3458), by a grant to S.O.R. from the German Ministry for Education and Research (13N15328/NG-FLIM), as well as by a grant to H.U. from the DFG (SFB1286/A8), and by a grant to T.S. (NIH grant R15NS095318).
Author information
Authors and Affiliations
Contributions
M.S.H. performed most of the fluorescence imaging experiments and the biochemistry experiments. T.M.D. developed software for data analysis and performed several fluorescence imaging experiments. S.J. also performed several fluorescence imaging experiments. T.S. performed all EM experiments (which were analyzed by V.S. and C.K.). S.M. and H.U. designed and performed all MS experiments. R.L. performed clustering analysis for Extended Data Fig. 3e,f. B.R. generated the synapse models. S.O.R. and M.S.H. designed the experiments and analyzed most of the data. All authors contributed to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review Information Nature Neuroscience thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
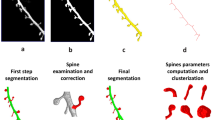
Extended Data Fig. 1 An integrative approach to analyze dendritic spines quantitatively.
We used serial-section electron microscopy (left panel) to determine the shapes, volumes and organelle distributions of the spines, and to generate the basic scaffold of 3D spines. We then used mass spectrometry to measure the neuronal protein copy numbers (top center panel), and we combined this with fluorescence microscopy, for every protein target, to obtain the copy numbers per spine. We also used super-resolution STED microscopy to determine the average location of the proteins, in tens of thousands of expert-annotated synapses (right panel). We then combined all of the information obtained with knowledge from more than 400 publications, to generate quantitative 3D models of average stubby and mushroom spines (bottom center panel).
Extended Data Fig. 2 Banker cultures are a good model for analyzing neuronal proteins.
a, Top rows: Banker cultures or neuron-glia co-cultures were immunostained for the neuronal marker β3-tubulin, for the GABAergic neuron marker glutamate decarboxylase (GAD65), for the glutamatergic neuron marker vGlut (vesicular glutamate transporter). Bottom rows: the cells were immunostained for the astrocyte marker glial fibrillary acidic protein (GFAP), for the microglia marker ionized calcium binding adaptor molecule 1 (Iba1), and for the oligodendrocyte marker oligodendrocyte transcription factor 2 (Olig2). Glutamatergic neurons represent the wide majority of the cells in the Banker cultures, with few other cell types present. Scale bar = 500 µm. b, An analysis of the protein copy numbers per cell, for different non-neuronal markers: myelin basic protein (MPB, myelin marker), glutamine synthetase (GS, astrocyte marker), GFAP, 10-Formyltetrahydrofolate Dehydrogenase (ALDH1L1, astrocyte marker), and myelin-oligodendrocyte glycoprotein (MOG, oligodendrocyte marker). The values were obtained in mass spectrometry analyses of Banker cultures (n = 4 experiments) or of rat brain homogenates (n = 8 experiments). The non-neuronal markers are virtually absent in Banker cultures. Whiskers show minimum and maximum values, the box shows quartiles and median.
Extended Data Fig. 3 Electron microscopy characterization of dendritic spines in hippocampal cultures, and Gauss shift clustering of spine morphological parameters.
a. Nine consecutive sections of a mushroom spine. Scale bar = 500 nm. b, Nine consecutive sections of a stubby spine. Scale bar = 500 nm. c, Average parameters obtained from 3D reconstructions of 30 mushroom spines and 34 stubby spines. The values show means ± SEM. d, The different panels show the total spine volume, spine surface area, PSD area, mitochondria volume, vacuole volume, and the number of small vesicles for mushroom and stubby spines. No significant differences were found (Kruskal-Wallis test followed by two-sided Dunn’s test). Thick lines indicate the median, dashed lines the upper and lower quartile. e, We also validated the findings from the EM dataset by comparing a large dataset of spines imaged with fluorescence microscopy. We relied on the lipid dye DiO to label the plasma membrane and to reveal the general morphology of the spines, while the PSD was indicated by an immunostaining for the marker Homer1. We then analyzed the area, height and width of the spine heads, along with the area occupied by the Homer1 signal, the intensity of this signal, and the size of an ellipse fitted on the Homer1 staining. Using these parameters, we could not find significant differences between the two types of spines, asit can be observed that curve of the morphology data is very similar to a 1 Gaussian cluster reference set, in contrast to a 3 Gaussian cluster reference set. Data shown in scale-space. The axes are transformed such that the graphs fit into the same coordinate system. The number of clusters is transformed such that “1.0” refers to the maximum number of clusters for the bandwidth segment we are interested in. The bandwidth is transformed such that the logarithm of the bandwidth lies between 0.0 and 1.0. f, Principal component analysis of the morphology data. No subpopulations can be identified. The bandwidth of the Gauss shift clustering is 0.5.
Extended Data Fig. 4 Post-hoc comparison of spine classification in this study to published criteria for automatic workflows.
We have checked our manual classifications of spines post-hoc, based on the criteria laid out in previous publications: (Risher, W. C., Ustunkaya, T., Alvarado, J. S. & Eroglu, C. Rapid golgi analysis method for efficient and unbiased classification of dendritic spines. PLoS One 9, (2014); Rodriguez, A., Ehlenberger, D. B., Dickstein, D. L., Hof, P. R. & Wearne, S. L. Automated three-dimensional detection and shape classification of dendritic spines from fluorescence microscopy images. PLoS One 3, (2008)), which define the threshold between mushroom and stubby spines as a length to width ratio of 1.05 (mean of criteria from these publications). In line with the published criteria, spines labeled as mushroom in this study exceed the defined length/width ratio (dotted line), whereas the vast majority of stubby spines are below it.
Extended Data Fig. 5 Detailed description of zone analysis for the STED images of spine proteins.
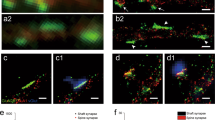
a, We separated the space occupied by spines in different zones, from the PSD area (red) to the dendritic shaft (green). The presence of different proteins in these zones was then analyzed. Scale bar = 500 nm. b, To generate 3D models of the spines, we projected the zones into the 3D space of two dendritic reconstructions. We chose two spines whose parameters (described in Extended Data Fig. 3) were closest to the average of all mushroom or, respectively, stubby spines investigated. The zones from panel (a) were then assigned in the model voxels, as shown in the graphs. The precise proportions of the zones, as in panel (a), were respected in the models. c, For modeling purposes, we also assigned different identities to the vacuoles present in these spines, as indicated in the color legend. Scale bar = 500 nm.
Extended Data Fig. 6 An analysis of nanomodules in the different spines.
a, We analyzed several bona fide PSD proteins, to determine their organization in nanoscale modules. Representative images of nanomodules are shown, the PSD is visualized in red-hot, the membrane in green. Arrows indicate modules, scale bar = 500 nm. The following proteins are shown: Mushroom 1 module – Homer1, 2 modules – Shank3, 3 modules – Shank3, >3 modules – Homer3; Stubby 1 module – Shank1, 2 modules – Shank1, 3 modules – PSD93, >3 modules – PSD95. b, As recently demonstrated15, the PSD is separated in two or more nanomodules for approximately 40% of the mushroom spines. Stubby spines showed the same pattern. Bar charts show mean ± SEM. Friedman test followed by two-sided Dunn’s tests. For information on sample size for each protein please refer to Supplementary Table 5.
Extended Data Fig. 7 Technical details on the biochemical analyses of Banker cultures, and an overview of the mass spectrometry results at the neuron level.
a, iBAQ mass spectrometry analysis (performed as in 28). The graphs indicate the correlation between results obtained in three different biological replicates (average R2 of 0.99, two-sided F-test). b-c, Dot blotting. For several proteins (Dopamine Receptor 1, Dopamine Receptor 2, LNGFR, mGluR2, Rapsyn, VAMP2) we employed dot blotting, as indicated in this example Neurogranin). Different amounts of purified proteins were blotted, in parallel with Banker cell lysates (panel a). The protein amounts were obtained by linear interpolation on the purified protein amounts (panel b). d, Over 6000 proteins were identified, in amounts spanning 8 orders of magnitude. e, An overview of the organelle distributions of the analyzed proteins. Cytosolic proteins make only one third of the proteins identified, in terms of protein species (identities), but sum to more than 60% of all proteins in terms of amounts.
Extended Data Fig. 8 Comparisons of our protein copy numbers with the literature, and examples of dendritic spines classified as “other”.
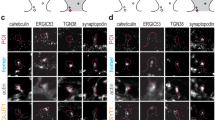
a, To validate our mass spectrometry analysis, we calculated the expected copy numbers for several proteins. First, we calculated the ratio of vATPase A and B subunit copy numbers to vATPase subunit a copies and compared it to its known stoichiometry (Kitagawa et al., 2008). 3 Second, we estimated the copy numbers per neuron of the presynaptic proteins Piccolo and Bassoon, based on their known copy numbers per synapse 2 and the known number of synapses in our preparation (see Extended Data Figure 9), and. we calculated the ratio against the respective measured quantities of Bassoon and Piccolo. Third, we compared the ratio of the core nucleosome subunits to H2A and the ratio of AP2 subunits α and β. The measured copy numbers are very close to the respective estimates (two-sided F-test; compare the linear fit, in red, with the identity line, in gray). b, A comparison of the copy numbers for 28 proteins (two-sided F-test) that had previously been estimated in the literature4 as well as: Antal, M. et al. Numbers, densities, and colocalization of AMPA- and NMDA-type glutamate receptors at individual synapses in the superficial spinal dorsal horn of rats. J. Neurosci. 28, 9692–701 (2008). Cheng, D. et al. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol. Cell. Proteomics 5, 1158–70 (2006). Lowenthal, M. S., Markey, S. P. & Dosemeci, A. Quantitative Mass Spectrometry Measurements Reveal Stoichiometry of Principal Postsynaptic Density Proteins. J. Proteome Res. 150415143859000 (2015) doi:10.1021/acs.jproteome.5b00109. Masugi-Tokita, M. et al. Number and density of AMPA receptors in individual synapses in the rat cerebellum as revealed by SDS-digested freeze-fracture replica labeling. J. Neurosci. 27, 2135–44 (2007). Nair, D. et al. Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. J. Neurosci. 33, 13204–24 (2013). Nusser, Z. et al. Cell type and pathway dependence of synaptic AMPA receptor number and variability in the hippocampus. Neuron 21, 545–559 (1998). Otmakhov, N. & Lisman, J. Measuring CaMKII concentration in dendritic spines. J. Neurosci. Methods 203, 106–114 (2012). Peng, J. et al. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry- Supplement. J. Biol. Chem. (2004). c, Comparisons for the same proteins (two-sided F-test), measured in three individual studies: Cheng, D. et al. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol. Cell. Proteomics 5, 1158–70 (2006). Peng, J. et al. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry- Supplement. J. Biol. Chem. (2004). Lowenthal, M. S., Markey, S. P. & Dosemeci, A. Quantitative Mass Spectrometry Measurements Reveal Stoichiometry of Principal Postsynaptic Density Proteins. J. Proteome Res. 150415143859000 (2015) doi:10.1021/acs.jproteome.5b00109. d-f, A comparison of the enrichment in the PSD of several proteins analyzed in STED microscopy in this study, and in Western Blot investigations of isolated PSDs in the study of 34. Panel (d) shows an analysis across all spines, irrespective of class; panel (e) shows mushroom spines, while panel (f) shows stubby spines. Overall, the biochemically-derived enrichment in the PSD correlates well with the enrichment observed by super-resolution microscopy. This relation is significant when mushroom spines are analyzed in isolation but is not significant for stubby spines (two-sided F-test). A possible conclusion is that stubby spines are less well controlled than mushroom spines (see also Fig. 5). g, During image analysis, we sometimes observed strong Homer1 (blue) staining without a clear mushroom or stubby shaped spine (membrane stained with DiO, green). These synapses were classified as “others”. Several representative images are shown. Note the diverse morphology. Scale bar = 500 nm.
Extended Data Fig. 9 An analysis of the number of synapses in cultured neurons.
a, Individual neurons were labeled fluorescently by overexpressing a membrane-bound GFP molecule (EGFP coupled to the palmitoylation domain of GAP43), which spreads to all neurites. Single neurons were then visualized (green). Two other color channels were used for immunostainings of synaptophysin (magenta), to reveal synaptic boutons, and for Ankyrin G (cyan), to reveal the axonal initial segments, and thereby to differentiate between dendrites and axonal branches. The boxed area is shown in panel c-d. b, Manual tracing of neurites, to identify all of the neurites of the labeled neuron (both axonal and dendritic). c, A high-zoom view, to enable the identification of the axon, labeled by Ankyrin G (cyan, arrowhead). The cyan signal to the lower left is due to staining in an adjoining neuron, not to staining in the green-labeled neuron. d, The same view, for the synaptophysin (magenta) channel. e, A higher-zoom view of the region boxed in panel d, which enables the visualization of synapses, consisting of synaptophysin-labeled boutons (magenta) closely apposed to GFP-labeled dendrites (green). Three examples are shown by arrowheads. Similarly, the number of presynapses was estimated by counting the number of synaptophysin-labeled boutons within the GFP-labeled axons. f, The number of pre- or postsynapses, shown as a box plot. The average values are 294 presynapses per neuron and 299 postsynapses per neuron, from 33 analyzed neurons, in 6 independent experiments. Box plots show the following: mid-line = median; boxes = 50th percentile; error bars = 75th percentile; symbols = outliers. Scale bars: 120 µm for panels a-b, 40 µm for panels c-d, 12 µm for panel e. g, Variation in the copy numbers of spine proteins. The graphs indicate the copy numbers of the proteins we analyzed in detail in the rest of the manuscript. The values are shown per neuron, from 4 independent experiments. Each dot show from one experiment, the lines indicate mean ± SEM.
Extended Data Fig. 10 Analyzing spine subclasses reveals no striking differences.
a, Mushroom spines were grouped into subclasses, based on head size, PSD size and neck length, using the median of the respective distributions as borders (red line). b, Scattering the different parameters against each other does not reveal obvious subpopulations. c, Stubby spines were grouped based on head and PSD size, again using the median of the respective distributions (red line). d, Scattering did not reveal obvious subpopulations. e, The percentages of spines in each subclass are shown. No subclasses show extremely high or low occupancy. f, We measured the intensity of each protein for each subclass, as a proxy of its abundance in the respective subclasses. We then divided the values by the overall intensity of the respective protein (averaged across all spines). This provides the relative abundance of each protein, in each spine subclass. The top panels show this analysis for the protein abundance in the whole volume of the spines, while the bottom panels show the analysis for the protein abundance in the PSD area. The dashed lines indicate the medians. We compared the protein abundance values between subclasses and found many significant differences. (Kruskal-Wallis followed by two-sided Dunn’s tests). For mushroom spines, the protein abundance across the whole spine is lower in spines exhibiting a long neck, and appears to be larger in spines with large PSDs. When only considering abundance in the PSD, the neck length no longer has a strong effect. The size of the PSD, as expected, correlates with protein abundance in the PSD. In stubby spines, a larger PSD also correlates with higher protein abundance, both in the whole spine volume and in the PSD. Head size does not affect protein abundance in either analysis. g, We then asked whether individual proteins tend to be significantly more abundant in particular spine subclasses. For this, we normalized protein abundances in each subclass by dividing the results from panel (f) by the median values of each subclass. As expected, this normalization removes any significant differences between the subclasses as a whole (Kruskal-Wallis followed by Dunn’s tests). However, now we could analyze each protein individually across subclasses, to test whether individual proteins are particularly abundant in the respective subclasses, independent of the overall effects induced by the morphology or the size of the respective subclasses. We found only extremely few differences. The results can be found in Supplementary Table 3 (two-sided Mann-Whitney U tests followed by Bonferroni correction).
Supplementary information
Supplementary Information
Supplementary Data 1 and 2.
Supplementary Table 1
A description of the materials used in the study.
Supplementary Table 2
A description of protein copy numbers in mushroom and stubby spines.
Supplementary Table 3
The statistical parameters for the analysis of spine subclasses, as shown in Extended Data Fig. 10.
Supplementary Table 4
A database of publications used in fine-tuning the models of spine morphology generated here.
Supplementary Table 5
The number of analyzed spines for each target for the experiments presented in Fig. 1 (excluding the homeostatic plasticity experiments).
Supplementary Table 6
An estimation of the total and individual protein amounts in spines of different types and sizes.
Supplementary Table 7
Calcium buffering capacity in spines, calculated according to the literature.
Supplementary Video 1
A movie of the organization of the average mushroom spine.
Supplementary Video 2
A movie of the organization of the average stubby spine.
Rights and permissions
About this article
Cite this article
Helm, M.S., Dankovich, T.M., Mandad, S. et al. A large-scale nanoscopy and biochemistry analysis of postsynaptic dendritic spines. Nat Neurosci 24, 1151–1162 (2021). https://doi.org/10.1038/s41593-021-00874-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-021-00874-w
This article is cited by
-
Preso enhances mGluR1-mediated excitotoxicity by modulating the phosphorylation of mGluR1-Homer1 complex and facilitating an ER stress after traumatic brain injury
Cell Death Discovery (2024)
-
Decreased CNNM2 expression in prefrontal cortex affects sensorimotor gating function, cognition, dendritic spine morphogenesis and risk of schizophrenia
Neuropsychopharmacology (2024)
-
Alternative splicing of latrophilin-3 controls synapse formation
Nature (2024)
-
HMGB1 mediates synaptic loss and cognitive impairment in an animal model of sepsis-associated encephalopathy
Journal of Neuroinflammation (2023)
-
Quantifying postsynaptic receptor dynamics: insights into synaptic function
Nature Reviews Neuroscience (2023)