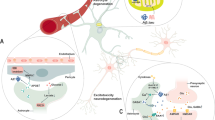
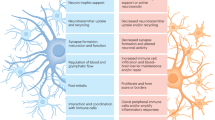
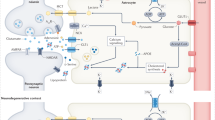
Abstract
The role of non-neuronal cells in Alzheimer’s disease progression has not been fully elucidated. Using single-nucleus RNA sequencing, we identified a population of disease-associated astrocytes in an Alzheimer’s disease mouse model. These disease-associated astrocytes appeared at early disease stages and increased in abundance with disease progression. We discovered that similar astrocytes appeared in aged wild-type mice and in aging human brains, suggesting their linkage to genetic and age-related factors.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Raw and processed mouse sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus database under accession number GSE143758 and are also available at https://singlecell.broadinstitute.org/single_cell/study/SCP302/mouse-alzheimers-and-disease-astrocytes. Source data for Fig. 1 are presented with the paper. Code is available at: https://github.com/naomihabiblab/5xFAD-sNucSeq.
References
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e17 (2017).
Strooper, B. & Karran, E. The cellular phase of Alzheimer’s disease. Cell 164, 603–615 (2016).
Baruch, K. et al. PD-1 immune checkpoint blockade reduces pathology and improves memory in mouse models of Alzheimer’s disease. Nat. Med. 22, 135–137 (2016).
Mathys, H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019).
Liddelow, S. A. et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017).
Zamanian, J. L. et al. Genomic analysis of reactive astrogliosis. J. Neurosci. 32, 6391–6410 (2012).
Matias, I., Morgado, J. & Gomes, F. Astrocyte heterogeneity: impact to brain aging and disease. Front. Aging Neurosci. 11, 59 (2019).
Rodríguez-Arellano, J., Parpura, V., Zorec, R. & Verkhratsky, A. Astrocytes in physiological aging and Alzheimer’s disease. Neuroscience 323, 170–182 (2016).
Bates, K., Fonte, J., Robertson, T., Martins, R. & Harvey, A. Chronic gliosis triggers Alzheimer’s disease-like processing of amyloid precursor protein. Neuroscience 113, 785–796 (2002).
Liao, M.-C. et al. Single-cell detection of secreted Aβ and sAPPα from human IPSC-derived neurons and astrocytes. J. Neurosci. 36, 1730–1746 (2016).
Habib, N. et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017).
Oakley, H. et al. Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 (2006).
Zeisel, A. et al. Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22 (2018).
Leitão, R. et al. Aquaporin-4 as a new target against methamphetamine-induced brain alterations: focus on the neurogliovascular unit and motivational behavior. Mol. Neurobiol. 55, 2056–2069 (2018).
Mucke, L. et al. Astroglial expression of human α1-antichymotrypsin enhances Alzheimer-like pathology in amyloid protein precursor transgenic mice. Am. J. Pathol. 157, 2003–2010 (2000).
Wang, C., Sun, B., Zhou, Y., Grubb, A. & Gan, L. Cathepsin B degrades amyloid-β in mice expressing wild-type human amyloid precursor protein. J. Biol. Chem. 287, 39834–39841 (2012).
Roussotte, F. F. et al. Combined effects of Alzheimer risk variants in the CLU and ApoE genes on ventricular expansion patterns in the elderly. J. Neurosci. 34, 6537–6545 (2014).
von und Halbach, O. Immunohistological markers for proliferative events, gliogenesis, and neurogenesis within the adult hippocampus. Cell Tissue Res. 345, 1–19 (2011).
Clarke, L. E. et al. Normal aging induces A1-like astrocyte reactivity. Proc. Natl Acad. Sci. USA 115, E1896–E1905 (2018).
Rothhammer, V. et al. Microglial control of astrocytes in response to microbial metabolites. Nature 557, 724–728 (2018).
Gaublomme, J. T. et al. Nuclei multiplexing with barcoded antibodies for single-nucleus genomics. Nat. Commun. 10, 2907–2908 (2019).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Haber, L. A. et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017).
Johnson, E. W., Li, C. & Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007).
McInnes, L., Healy, J. & Melville, J. UMAP: Uniform Manifold Approximation and Projection for dimension reduction. Preprint available at arXiv https://arxiv.org/abs/1802.03426 (2018).
Lancichinetti, A. & Fortunato, S. Benchmarks for testing community detection algorithms on directed and weighted graphs with overlapping communities. Phys. Rev. E 80, 016118 (2009).
Habib, N. et al. Div-Seq: single-nucleus RNA-seq reveals dynamics of rare adult newborn neurons. Science 353, 925–928 (2016).
Angerer, P. et al. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243 (2016).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019).
Liberzon, A. et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Smillie, C. S. et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178, 714–730.e22 (2019).
Szklarczyk, D. et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Munkres, J. Algorithms for the assignment and transportation problems. J. Soc. Ind. Appl. Math. 5, 32–38 (1957).
Baruch, K. et al. Breaking immune tolerance by targeting Foxp3+ regulatory T cells mitigates Alzheimer’s disease pathology. Nat. Commun. 6, 7967–12 (2015).
Acknowledgements
We thank R. Herbst, I. Avraham-Davidi and members of the Habib laboratory for discussions and helpful suggestions, and L. Gaffney for graphics and J. Rood for editing. We thank I. Shif and M. Bronstein for help with library preparation and sequencing. This work was supported by the NIH BRAIN Initiative grant (grant no. U19MH114821 for A.R.), the Klarman Incubator and Klarman Cell Observatory (A.R.), the HHMI (A.R.) and a MOST-IL flagship grant (for N.H.). M.S. holds the Maurice and Ilse Katz Professorial Chair in Neuroimmunology, and N.H. holds the Goren-Khazzam Chair in Neurobiology. The work was supported in part by Advanced European Research Council grant no. 232835 (M.S.); Israel Science Foundation (ISF) research grants no. 991/16 (M.S.), no. 913/15 (T.K.) and no. 1250/18 (T.K.); ISF-Legacy Heritage Biomedical Science Partnership research grant no. 1354/15 (M.S.); the Adelis Foundation and the Thompson Foundation (M.S.); the MOST-IL-China research grant no. 3-15687 (N.H.); and the Alon Fellowship (N.H.).
Author information
Authors and Affiliations
Contributions
N.H., M.S., A.R. and F.Z. conceived the study. M.S., N.H. and S.M. designed the experiments. C.M. and D.K. isolated nuclei and generated libraries with assistance from D.D. and L.N. S.M., J.L.M. and F.C. conducted the imaging experiments and S.M. analyzed the imaging data. S.M. and R.D.-S. conducted the animal work. N.H., A.R. and T.K. devised the data analysis. N.H., M.V., G.G. and T.K. conducted the data analysis. T.K. and N.H. developed a new algorithmic approach. M.S., N.H., T.K. and A.R. wrote the paper with input from all of the authors.
Corresponding authors
Ethics declarations
Competing interests
A.R. is a founder and equity holder of Celsius Therapeutics, an equity holder in Immunitas Therapeutics and an SAB member of Syros Pharmaceuticals, Thermo Fisher Scientific, Neogene Therapeutics and Asimov. The other authors declare no competing interests.
Additional information
Peer review information Nature Neuroscience thanks Valentina Fossati and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 A cellular map of the mouse hippocampus of WT and 5xFAD mice and quality controls.
a, Doublet detection and elimination. 2-D tSNE embedding of 60,818 single nuclei RNA profiles from hippocampus of four WT and four 5xFAD 7- month old mice, before filtration. Top: Color coded by cluster assignment. Bottom: Color coded by doublet score assigned per cell by the Scrublet29 software, used to infer doublet cells and clusters to exclude from the analysis. b, Number of genes and transcripts across clusters. Violin plots showing the distribution of number of genes (top) and transcripts (unique UMIs, bottom) detected in each cluster (n=8 mice, 10 samples). Cluster numbers as in Fig. 1b. c, 2-D tSNE embedding of single nuclei RNA profiles from hippocampus of WT and 5xFAD mice (as in Fig. 1b), colored by (from left to right): batch, mouse strain (WT or AD) and sample. d, Similar distribution of samples and batches across clusters. The percent of cells per cluster, in WT and 5xFAD mice. Middle: Colored by batch/lysis buffer (red= EZ lysis. Blue = NP40 lysis, Methods), Right: Colored by sample (blue color scale, 4 animals and 5 samples per mouse strain, AD or WT). Left: The hierarchical cluster tree and annotations of clusters, as in Fig. 1b.
Extended Data Fig. 2 Cell type marker genes and assignments.
a, Expression of marker genes across clusters. 2-D tSNE embedding of single nuclei RNA profiles from hippocampi of WT and 5xFAD mice (as in Fig. 1b), colored by expression levels of marker genes: Grin2b (neurons), Gad2 (GABAergic neurons), Vcan (Oligodendrocytes precursor cells, OPCs), Hmha1 (microglia), Flt1 (endothelial), Vtn (pericytes), Plp1 (oligodendrocytes), Slc1a3 (astrocytes), Gfap (astrocytes), Rarres2 (ependymal/NPCs), Slc6a13 (fibroblasts), Homer1 (immediate early gene, IEGs). b, Clusters and marker genes. Dot plot showing the expression level (color scale) and the percent of cells expressing (dot size) marker genes across all clusters (rows). Cluster numbers as in Fig. 1b. c, Disease associated microglia (DAM) signature enriched in AD. Violin plots showing the distribution in WT (n=8 animals, 896 cells) and AD (n=8 animals, 1,540 cells) of microglia expression scores for signatures of genes up-regulated in DAM compared to homeostatic microglia (from keren-Shaul et al.1, Methods). Expression score per cell is the geometric mean normalized expression level (TPMs) across all signature genes, corrected by subtraction of the geometric mean expression of a random set of genes of similar expression levels (Methods). d, Recently activated pyramidal neurons. Left: Dot plot as in (b) showing the expression of immediate early genes (IEGs) across all clusters (as in Fig. 1b), showing cluster 23, capturing pyramidal neurons expressing IEGs. Right: tSNE plot of all cells, color coded by the expression level of the Egr4 gene in CA3/CA1/Subiculum (cluster 23), and DG excitatory neurons (part of cluster 12). e, Cell type specific markers. Dot plots as in (b) showing the expression level, across all clusters (rows, as in Fig. 1b), of markers found to be specific to cells classified as (from left to right): ependymal/NPCs, fibroblasts and pericytes.
Extended Data Fig. 3 Diversity of astrocyte states in WT and 5xFAD mice.
a, 2-D Umap25 embedding of 7,345 single nuclei RNA profiles of astrocytes (as in Fig. 1d) from hippocampus of 4 WT (left) and 4 5xFAD (AD, right) 7-month old mice. Colored by cluster, all other cells in light yellow in the background. b, 2-D Umap25 embedding as in (a), colored by sample (left), or batch (right). c, Number of genes and transcripts across clusters. Violin plots showing the distribution of number of genes (top) and transcripts (unique UMIs, bottom) detected per cluster (n=8 animals, total of 7,345 cells). Cluster numbers as in (a). d, Diffusion maps28 of 7,345 single nuclei RNA profiles of astrocytes in the hippocampus of WT and 5xFAD mice, showing 2-D embedding of cells in combinations of the top four diffusion components (DC), colored by mouse strain, WT (blue) and 5xFAD (AD, red). e, Distribution of astrocyte states in WT and AD brains. Box plots showing the fraction of each astrocyte cluster (compared to total number of astrocyte cells, clusters as in Fig. 1d, n=8 animals, 10 samples), in WT and 5xFAD mice. Displaying the median (thick lines), 25% and 75% quantiles (box), and individual samples (dots).
Extended Data Fig. 4 DAAs found in female 5xFAD mice and in the cortex.
a, tSNE embedding of single nuclei RNA profiles of astrocytes from hippocampus of 7-month old female (1,500 nuclei, 2 mice, left) and male (5,183 nuclei, 4 mice, right) of three WT and three 5xFAD (AD) mice. Colored by cluster assignment. The three astrocyte end states are marked on the graph (annotated by projection from Fig. 1d, Methods). b, tSNE embedding as in (a), colored by cluster 4 (DAA) and cluster 6 (Gfap-low) cluster IDs of male mice as in Fig. 1d (other cells in grey). c, Expression levels (color scale) and the percent of cells expressing (dot size) of marker genes across clusters, split by sex: for DAA (Ggta1, Gsn, Osmr, Vim, Serpina3n, Ctsb, Gfap), Gfap-low (Fabp7, Slc38a1, Myoc, Aqp4, Id1, Id3, Gfap) and Gfap-low (Mfge8, Slc7a10, Luzp2). d, The proportion of astrocytes classified as Gfap-low (clusters 1,2,5,6 in (a)), DAAs (cluster 3 in (a)) and Gfap-high (cluster 4 in (a)) in male and female mice. Bar: individual mice, colored by strain and sex. e, DAAs in 5xFAD mice cortex at age 7 and 10 months. tSNE embedding of single nuclei RNA profiles of astrocytes from WT and AD mice, from the cortex of 7 and 10 month old mice (6,062 nuclei, 4 mice) and hippocampus of 7-month old mice (5,344 nuclei, 4 mice). Colored by cluster. f, Similar astrocytes marker genes in in the hippocampus and cortex. Dot plot showing the expression level (as color scale) and the percent of cells expressing (as dot size) marker genes for DAAs, Gfap-high and Gfap-low across clusters (as in (e)), split by brain region to hippocampus and cortex. g, tSNE embedding as in (e), colored by predicted cluster ID of hippocampal astrocytes from 7-month old male mice (inferred by CCA22,33 projections, as in Fig. 1d, Methods). h, Cortical astrocyte populations match astrocyte states identified in the hippocampus. Heat map showing the correspondence between the de novo cluster IDs (rows, from (e)) of the cross regional dataset, and the predicted cluster IDs (columns) using the hippocampal astrocytes cluster IDs as reference (as from Fig. 1d). Color scale based on the proportion of predicted IDs per de novo cross-regional cluster. i, DAAs appear in the cortex of 5xFAD (AD) mice. The proportion of astrocytes, per sample, across clusters, including clusters of Gfap-low, DAAs, and Gfap-high astrocytes. Bars: Individual mice, color annotated by region: cortex or hippocampus (Hip), age: 7 or 10 months (m), and strain: AD or WT. j, Astrocyte marker genes. Left: Average expression level (color scale) and the percent of cells expressing (dot size) marker genes for: Gfap-low (Slc7a10; Trpm3), DAA (Ctsb16; Csmd1, associated with cognitive functions; C4b, encoding complement factor 4; Vim, a marker of adult neurogenesis/NSCs18), common to DAA and Gfap-high (Cd9, expressed by neural stem cells like astrocytes), and Gfap-high (Sparcl1/Hevin, encoding astrocytes pro-synaptic protein; Aqp4, an endfeet marker 14) (cells clusters as in Fig. 1e). Right: Violin plots (n=8 animals, 10 samples, 7,345 cells), showing the expression level distributions of Csmd1 and Ctsb in WT and AD astrocytes.
Extended Data Fig. 5 Shared and distinct transcriptional programs of DAAs and physiological Gfap-high astrocytes.
a, Differential expression across astrocyte states. Volcano plots showing differentially expressed genes in each pair of states (n=8 animals, 10 samples. y-axis: -log adjusted hypergeometric p-value, following FDR multiple hypothesis correction, x-axis: average log fold change). AD risk factor genes from GWAS marked in orange (as in Fig. 2a). b, DAAs, Gfap-high astrocytes, and cluster 3 astrocytes share multiple upregulated genes and pathways compared to the homeostatic Gfap-low astrocyte population, but also have distinct expression programs. Pathway (rows) enrichment for upregulated genes in cluster 4 (C4, DAAs, n=478 cells), cluster 6 (C6, Gfap-high, n=457), or cluster 3 (C3, intermediate state, n=1,666 cells), compared to Gfap-low astrocytes (n=1,594 cells). Enriched pathways30 (hypergeometric p-value with FDR<0.05. n=8 animals, 10 samples), colored by -log FDR values (as in Fig. 2b, with full list of pathway annotations and no scaling).
Extended Data Fig. 6 DAAs express signatures of reactive astrocytes and are found across brain regions.
a-b, Signatures of inflammatory reactive astrocytes found in AD. Ridge plots showing the distribution of expression scores across each astrocyte cluster, for signature genes of: pan-reactive, A1, and A2 (from Liddelow et al.5, left), or of: pan-reactive, inflammation (LPS-induced), and ischemia (MCAO-induced) (from Zamanian et al6. right). Expression score per cell is the geometric mean normalized expression level (TPMs) across all signature genes, corrected by subtraction of the geometric mean expression of a random set of genes of similar expression levels. b, Violin plots showing the distribution of expression scores across WT (3,831 cells, n=4 animals) and AD (3,514 cells, n=4 animals), for signature genes for: A1 and A2 astrocytes (as defined in Liddelow et al.5). Scores computed as in (a). c, Expression of genes from reactive, A1, and A2 signatures, showing diversity of gene patterns across astrocyte clusters. 2-D umap25 embedding of 7,345 single nuclei RNA profiles of astrocytes in the hippocampus of WT and 5xFAD mice (as in Supplementary Fig. 3a), colored by the gene expression level. Dotted black lines: areas of highest expression. Gene name: on top of each graph. The associated gene (pan reactive, A1 or A2) on the right side of each panel. d, Astrocytes expressing DAA markers are present in AD brains, enriched in the subiculum and in proximity to Aβ plaques. Representative immunofluorescence images (staining repeated over n=4 AD and WT mice, with 4 brain slices per animal) in sagittal sections of 7-8-month old 5XFAD mice (with Fig. 2f-h). From top to Bottom: Subiculum, stained for GFAP (green), VIM (red), and serpinA3N (gray), (as in Fig. 2f). Dentate gyrus (in Fig. 2g, left) and subiculum (in Fig. 2g, right), stained for GFAP (green), VIM (red), and Aβ (gray). Subiculum, stained for GFAP (green), serpinA3N (red), and Aβ (gray) (as in Fig. 2h). Cell nuclei are shown in blue (Hoechst). Scale bar, 50μm.
Extended Data Fig. 7 Clustering of astrocyte cells from WT and 5xFAD mice across ages.
a-b, tSNE embedding of sNuc-Seq profiles of astrocytes in WT and 5xFAD (AD) mice across 6 different age groups (in months): 1.5-2 (n=6 mice), 4-5 (n=4), 7-8 (n=8), 10 (n=2), 13-14 (n=6), and 20 (n=2, WT only). (23,863 cells, integrated across four batches22,33, Methods). Colored by: cluster IDs (in (a)), or predicted cluster IDs (using reference cluster IDs of astrocytes of 7-month old mice as in Fig. 1d, in (b)). c, The correspondence between the de novo cluster IDs (columns) of the time course data, and the predicted cluster IDs (rows) from astrocytes of 7-month old mice (as in Fig. 1d). Color scale = proportion of predicted IDs per de novo cluster. d-g, 2-D tSNE embedding of astrocytes across ages as in (a), colored by: age (d), mouse strain, AD/WT (e), batch (f), or sample (g). h, Continuous trajectory of astrocyte states across ages in WT and AD. Diffusion map embedding of astrocytes across ages. Colored by de novo clusters: Gfap-low (1), Gfap-low/intermediate (2), Gfap-high (3), and DAAs (4). 15,113 astrocyte cells, down sampled randomly from 23,863 cells to capture 2,500 cells of each age group (or the maximum number of cells available if less than 2,500). i, Diffusion map embedding (as in (h)), colored by predicted cluster IDs (as in (b)). j, 2-D embedding of the 15,113 astrocytes of AD (left) and WT (right) cells across ages, projected onto the diffusion map of 7-month old mice (by the weighted average position of the K-NN from Fig. 1d, Methods). Colored by the de novo clusters (as in (a)). k, The expression level (as color scale) and the percent of cells expressing (as dot size) of marker genes for DAAs (Ggta1, Gsn, Osmr, Vim, Serpina3n, Ctsb, and Gfap), Gfap-high (Fabp7, Slc38a1, Myoc, Aqp4, Id1, Id3, and Gfap) and Gfap-low (Mfge8, Slc7a10, Luzp2), across de novo clusters of astrocytes across ages.
Extended Data Fig. 8 DAAs are largely derived from Gfap-low astrocytes.
a-b, Direction of transition of AD astrocytes from their global optimal nearest neighbor (origin) cell (predicted by the Hungarian algorithm34, Methods) to their given position in the diffusion map. For each cluster, force field (black arrows) marking the directionality of transitions along the diffusion map28 (as in Fig. 1d), cells colored by cluster ID. Showing transitions from the predicted cell of origin among all WT cells or AD cells outside the cluster (a), or among all WT cells only (b) (transitions among AD cells only are shown in Fig. 3). c, Proportion of cells of origin per cluster (dot size and color) for each AD cluster (rows), from all WT cells (left) or from AD cells from all other clusters (right). d, Scheme of predicted transitions, shown as a graph with arrows between pairs of clusters with high proportion of origin cells (>15%) when mapping AD to all WT cells. Color and width of edge reflects the proportion. Diffusion map as in (a) in the background. e, Genes correlated with predicted transitions from WT cells to DAA in AD. The expression level across clusters (dot color) and the percent of cells expressing (dot size) significant (Pearson Correlation coefficient, FDR q-value<0.001, n=28 mice, 25,076 cells, Supplementary Table 1) genes that correlated with the transition to cluster 3 (intermediate) or cluster 4 (DAAs) from WT cells. Bottom: Assignment of each gene to a transition between pairs of clusters. f, Direction of transition on the diffusion space of AD astrocytes in each cluster from their weighted K-nearest neighbor position among all other WT cells. Force field showing the directionality of transitions between the expected position in the diffusion map (weighted average position of K-NN among all WT cells, k=10) to the true position along the diffusion map (as in Fig. 1d) for each cell. Colored by cluster IDs. Cluster numbers on top of each graph.
Extended Data Fig. 9 DAA-like astrocytes found in aging human cortex.
a-b, Diversity of human astrocytes. De novo tSNE embedding of 3,392 sNuc-Seq profiles of cortical human astrocytes from post-mortem aging brains of AD and non-AD individuals (n=48, from Mathys et al.4), colored by de novo human cluster ID (a) or by the predicted mouse cluster IDs (b) (predicted by CCA22,33 projections from the mouse clusters as in Fig. 1d). c, The average prediction scores (as color scale) and the percent of cells with score above 0 (as dot size) for the three major end-states mouse clusters (from Fig. 1d), across the human astrocyte clusters (as in a, Mathys et al.4). From left to right: Gfap-low (mouse cluster 1), DAAs (mouse cluster 4), and Gfap-high (mouse cluster 6). d, Dot plot showing the expression level (as color scale) and the percent of cells expressing (as dot size) marker genes for mouse astrocyte states: DAA (OSMR, VIM, GFAP), Gfap-high (ID1, ID3, SLC38A1, GFAP) and homeostatic Gfap-low (MFGE8), across clusters of the human cortical astrocytes (from Mathys et al.4).
Supplementary information
Supplementary Tables
Supplementary Tables 1–4
Source data
Source Data Fig. 1
Source Data with: t-SNE coordinates, cluster ID, mouse strain, sample ID, per cell.
Rights and permissions
About this article
Cite this article
Habib, N., McCabe, C., Medina, S. et al. Disease-associated astrocytes in Alzheimer’s disease and aging. Nat Neurosci 23, 701–706 (2020). https://doi.org/10.1038/s41593-020-0624-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-020-0624-8
This article is cited by
-
Blockage of VEGF function by bevacizumab alleviates early-stage cerebrovascular dysfunction and improves cognitive function in a mouse model of Alzheimer’s disease
Translational Neurodegeneration (2024)
-
The interaction between ageing and Alzheimer's disease: insights from the hallmarks of ageing
Translational Neurodegeneration (2024)
-
Neuropathogenesis-on-chips for neurodegenerative diseases
Nature Communications (2024)
-
Pathological phenotypes of astrocytes in Alzheimer’s disease
Experimental & Molecular Medicine (2024)
-
Longitudinal markers of cerebral amyloid angiopathy and related inflammation in rTg-DI rats
Scientific Reports (2024)