Abstract
Mammalian circadian behaviors are orchestrated by the suprachiasmatic nucleus (SCN) in the ventral hypothalamus, but the number of SCN cell types and their functional roles remain unclear. We have used single-cell RNA-sequencing to identify the basic cell types in the mouse SCN and to characterize their circadian and light-induced gene expression patterns. We identified eight major cell types, with each type displaying a specific pattern of circadian gene expression. Five SCN neuronal subtypes, each with specific combinations of markers, differ in their spatial distribution, circadian rhythmicity and light responsiveness. Through a complete three-dimensional reconstruction of the mouse SCN at single-cell resolution, we obtained a standardized SCN atlas containing the spatial distribution of these subtypes and gene expression. Furthermore, we observed heterogeneous circadian gene expression between SCN neuron subtypes. Such a spatiotemporal pattern of gene regulation within the SCN may have an important function in the circadian pacemaker.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jun Yan (junyan@ion.ac.cn). Cell body distributions, reconstructed spatial domains and spatial gene expression from LCM-seq data can be accessed from our website: http://yanlab.org.cn/scn-atlas. The raw data files and DGEs for both Drop-seq and LCM datasets reported in this article have been deposited in the Gene Expression Omnibus database under GSE117295, GSE118403 and GSE132608.
References
Hastings, M. H., Maywood, E. S. & Brancaccio, M. Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 19, 453–469 (2018).
Takahashi, J. S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017).
Yamaguchi, S. et al. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302, 1408–1412 (2003).
Schwartz, W. J., Gross, R. A. & Morton, M. T. The suprachiasmatic nuclei contain a tetrodotoxin-resistant circadian pacemaker. Proc. Natl Acad. Sci. USA 84, 1694–1698 (1987).
Maywood, E. S. et al. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr. Biol. 16, 599–605 (2006).
Lee, I. T. et al. Neuromedin S-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron 85, 1086–1102 (2015).
Abrahamson, E. E. & Moore, R. Y. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 916, 172–191 (2001).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Chen, R., Wu, X., Jiang, L. & Zhang, Y. Single-cell RNA-Seq reveals hypothalamic cell diversity. Cell Rep. 18, 3227–3241 (2017).
Hughes, M. E., Hogenesch, J. B. & Kornacker, K. JTK-CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms 25, 372–380 (2010).
Hatori, M. et al. Lhx1 maintains synchrony among circadian oscillator neurons of the SCN. eLife 3, 1–16 (2014).
Schnell, A. et al. The nuclear receptor REV-ERBα regulates Fabp7 and modulates adult hippocampal neurogenesis. PLoS ONE 9, e99883 (2014).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Cheng, A. H. et al. SOX2-dependent transcription in clock neurons promotes the robustness of the central circadian pacemaker. Cell Rep. 26, 3191–3202.e8 (2019).
Romanov, R. A. et al. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat. Neurosci. 20, 176–188 (2017).
Park, J. et al. Single-cell transcriptional analysis reveals novel neuronal phenotypes and interaction networks involved in the central circadian clock. Front. Neurosci. 10, 481 (2016).
Crow, M., Paul, A., Ballouz, S., Huang, Z. J. & Gillis, J. Characterizing the replicability of cell types defined by single cell RNA-sequencing data using MetaNeighbor. Nat. Commun. 9, 884 (2018).
Paul, A. et al. Transcriptional architecture of synaptic communication delineates GABAergic neuron identity. Cell 171, 522–539.e20 (2017).
Bindea, G. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009).
Berson, D. M., Dunn, F. A. & Takao, M. Phototransduction by retinal ganglion cells that set the circadian clock. Science 295, 1070–1073 (2002).
Hattar, S., Liao, H. W., Takao, M., Berson, D. M. & Yau, K. W. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science 295, 1065–1070 (2002).
Karatsoreos, I. N., Wang, A., Sasanian, J. & Silver, R. A role for androgens in regulating circadian behavior and the suprachiasmatic nucleus. Endocrinology 148, 5487–5495 (2007).
Hannibal, J., Hundahl, C., Fahrenkrug, J., Rehfeld, J. F. & Friis-Hansen, L. Cholecystokinin (CCK)-expressing neurons in the suprachiasmatic nucleus: innervation, light responsiveness and entrainment in CCK-deficient mice. Eur. J. Neurosci. 32, 1006–1017 (2010).
Morin, L. P. SCN organization reconsidered. J. Biol. Rhythms 22, 3–13 (2007).
Brancaccio, M. et al. Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science 363, 187–192 (2019).
Brancaccio, M., Patton, A. P., Chesham, J. E., Maywood, E. S. & Hastings, M. H. Astrocytes control circadian timekeeping in the suprachiasmatic nucleus via glutamatergic signaling. Neuron 93, 1420–1435.e5 (2017).
Tso, C. F. et al. Astrocytes regulate daily rhythms in the suprachiasmatic nucleus and behavior. Curr. Biol. 27, 1055–1061 (2017).
Barca-Mayo, O. et al. Astrocyte deletion of Bmal1 alters daily locomotor activity and cognitive functions via GABA signalling. Nat. Commun. 8, 14336 (2017).
Mazuski, C. et al. Entrainment of circadian rhythms depends on firing rates and neuropeptide release of VIP SCN neurons. Neuron 99, 555–563.e5 (2018).
Inagaki, N., Honma, S., Ono, D., Tanahashi, Y. & Honma, K. I. Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc. Natl Acad. Sci. USA 104, 7664–7669 (2007).
Fernandez, D. C., Chang, Y.-T., Hattar, S. & Chen, S.-K. Architecture of retinal projections to the central circadian pacemaker. Proc. Natl Acad. Sci. USA 113, 6047–6052 (2016).
Mieda, M. et al. Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron 85, 1103–1116 (2015).
Fukuda, H., Tokuda, I., Hashimoto, S. & Hayasaka, N. Quantitative analysis of phase wave of gene expression in the mammalian central circadian clock network. PLoS ONE 6, e23568 (2011).
Welsh, D. K., Takahashi, J. S. & Kay, S. A. Suprachiasmatic nucleus: cell autonomy and network properties. Annu. Rev. Physiol. 72, 551–577 (2010).
Hamada, T., Antle, M. C. & Silver, R. Temporal and spatial expression patterns of canonical clock genes and clock-controlled genes in the suprachiasmatic nucleus. Eur. J. Neurosci. 19, 1741–1748 (2004).
Gonze, D., Bernard, S., Waltermann, C., Kramer, A. & Herzel, H. Spontaneous synchronization of coupled circadian oscillators. Biophys. J. 89, 120–129 (2005).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Alles, J. et al. Cell fixation and preservation for droplet-based single-cell transcriptomics. BMC Biol. 15, 44 (2017).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Hou, B. et al. Scalable and DiI-compatible optical clearance of the mammalian brain. Front. Neuroanat. 9, 19 (2015).
Tomer, R., Ye, L., Hsueh, B. & Deisseroth, K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 9, 1682–1697 (2014).
Waltman, L. & Van Eck, N. J. A smart local moving algorithm for large-scale modularity-based community detection. Eur. Phys. J. B 86, 471 (2013).
García-Alonso, C. R., Pérez-Naranjo, L. M. & Fernández-Caballero, J. C. Multiobjective evolutionary algorithms to identify highly autocorrelated areas: the case of spatial distribution in financially compromised farms. Ann. Oper. Res. 219, 187–202 (2014).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Wu, G., Anafi, R. C., Hughes, M. E., Kornacker, K. & Hogenesch, J. B. MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics 32, 3351–3353 (2016).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Feng, L., Zhao, T. & Kim, J. neuTube 1.0: A new design for efficient neuron reconstruction software based on the SWC format. eNeuro 2, https://doi.org/10.1523/ENEURO.0049-14.2014 (2015).
Pettersen, E. F. et al. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Yan, J., Wang, H., Liu, Y. & Shao, C. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput. Biol. 4, e1000193 (2008).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Acknowledgements
This work was supported by grants from the Natural Science Foundation of China (grant no. 31571209 to J.Y.), the Strategic Priority Research Program of Chinese Academy of Sciences (grant no. XDB32040100 to J.Y.), the Shanghai Municipal Science and Technology Major Project (grant no. 2018SHZDZX05 to J.Y.), the Natural Science Foundation of Shanghai (grant no. 16ZR1448800 to H.W.) and the National Science Foundation for Young Scientists of China (grant no. 31701029 to H.W.). We thank M.-m. Poo (Institute of Neuroscience, Chinese Academy of Sciences) for reading the manuscript and helpful discussions.
Author information
Authors and Affiliations
Contributions
J.Y., S.W., D.M., M.Z. and L.X. produced the experimental design. S.W., L.X. and C. Z. did the Drop-seq analysis. D.M. and Q.W. did the smFISH analysis. Q.W. and L.X. did the tissue clearing and imaging. M.Z. did the LCM RNA-seq. Y.F. provided validation and husbandry of the mouse lines. D.M. constructed the 3D SCN atlas. L.G. implemented the website. S.W., D.M., M.Z., L.X. and Q.W. performed experiments and data acquisition. S.W., J.Y., D.M., H.W. and L.G. performed the data analysis, and interpretation and generation of figures. J.Y., S.W., D.M. and H.W. wrote, reviewed and edited the manuscript. J.Y. and H.W. dealt with scientific direction and funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Neuroscience thanks S. Panda, H. Ueda, Z. Yao and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Comparisons of our SCN Drop-seq data with previously published datasets and circadian gene expression profiles of common rhythmic genes across SCN basic cell types.
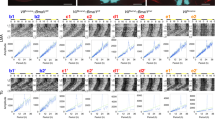
a, Comparison of cell clusters between our data and Chen et al.’s single-cell RNAseq data in mouse hypothalamus. The similarities of cell clusters in two datasets were represented by the coefficients of non-negative linear regression analysis. b, Percentage of cells in “projection set” that can be consistently projected to specific cell types or SCN neuron subtypes when projecting to entire “training set” and a subset of “training set”. The training set was randomly divided into 6 subsets (n=6). The boxplots indicate the median, minimum, maximum, first quartile and third quartile of the distribution of percentages. c, t-SNE plot showing the distributions of cells from different batches. d, Cell compositions of different cell types (left) and SCN neuron subtypes (right) in samples collected at different time points. e, Heatmap showing the expression profiles of top 10 marker genes across all cell types. f, Comparison of circadian phases of 120 rhythmic genes in our data with those from a published circadian transcriptomic dataset of bulk SCN from Hatori et al.’s study. Rhythmic genes in our data and their phases used here were obtained by pooling all cells at the same circadian time point. Dash lines indicate the ± 2 hours of phase difference. Pearson’s correlation coefficient and p-value were determined by cor.circular test in R package circular. g, Circadian gene expression profiles of 26 genes that are rhythmically expressed in more than 4 basic cell types across all cell types in SCN (core circadian genes, in blue; protein folding related genes, in green). Genes are ordered by their circadian phases in Neurons. Colors represent scaled gene expression values across all cell types at different circadian time points.
Extended Data Fig. 2 Expression profiles of common rhythmic genes and around-the-clock GO analysis in different cell types.
a, smFISH result showing circadian Bmal1 expression of Vip+ neurons (top) and Avp+ neurons (middle) in SCN and ependymal cells lining the third ventricle (bottom). DAPI signals in blue color indicate the positions of cell bodies. Scale bars represent 10μm. b, Quantification of circadian Bmal1 expression in (A). The boxplots indicate the median, minimum, maximum, first quartile and third quartile of the distribution of single-cell Bmal1 expression. Circadian phase (φ) and its error were analyzed by cosine regression on the population median values of each subtype at each time point. Circadian phase (φ) of Bmal1 is at CT18 in Avp+ neurons and Vip+ neurons but at CT24 in ependymal cells. The numbers of Vip neurons used in the analysis: n=265 (CT14); n=252 (CT18); n=169 (CT22); n=257 (CT26); n=275 (CT30); n=250 (CT34). The numbers of Avp neurons used in the analysis: n=444 (CT14); n=423 (CT18); n=283 (CT22); n=431 (CT26); n=462 (CT30); n=420 (CT34). The numbers of ependymal cells used in the analysis: n=51 (CT14); n=62 (CT18); n=73 (CT22); n=52 (CT26); n=67 (CT30); n=67 (CT34). P-values of comparisons between peak and trough values using two-sided Mann-Whitney U-test are p=2.4x10-20 (Vip+), p=1.2x10-7 (Avp+), and p=3.0x10-19 (ependymal cells). P-values are represented as * p<0.05, ** p<0.01, *** p<0.001. (c) t-SNE plot showing the clustering result of neurons without filtering the circadian effect. Note that cluster 5 only consists of cells collected in subjective day. d, Go terms enriched in specific time windows around the clock among rhythmic genes of 6 major SCN cell types including endothelial cells, microglia, neurons, astrocytes, NG2 cells and oligodendrocytes. The significance of enrichment is represented by -log(p-value) and scaled by row. The result of ependymal cells is shown in Fig. 2f. P-values were calculated with Fisher’s exact test.
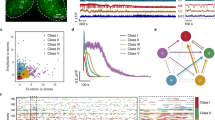
Extended Data Fig. 3 Classification of neurons in and around SCN from our Drop-seq data.
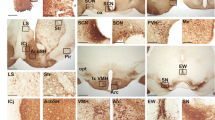
a, Flow chart of our iterative procedure to remove circadian effect on neuron classification. b, Violin plot of selected marker genes in each neuron cluster. The dendrogram of all 16 neuron subtypes (left) are constructed based on the Euclidean distances between the expression patterns of their marker genes (see also in Fig. 3c). c, Comparison of neuronal clusters between our data and Chen et al.’s (top) or Romanov et al.’s (bottom) single-cell RNAseq data in mouse hypothalamus. The similarities of neuronal clusters in two datasets were represented by the coefficients of non-negative linear regression analysis. d, Enrichment of the regulon in each neuron cluster. The regulons shown here are the same as in Fig. 3f but showing all SCN and non-SCN neuron clusters. Expression level of each regulon was normalized to z-score by row. e, ISH images of 6 SCN enriched genes (upper) and 6 SCN depleted genes (lower) were from Allen Brain Atlas.
Extended Data Fig. 4 Analysis of SCN neuron subtypes and comparison between Drop-seq data with 10X data.
a, Pie chart showing the composition of SCN neuron subtypes in our Drop-seq data. b, Clustering tree of SCN 10X dataset. Avp+/ Nms+, Vip+/ Nms+ and Grp+/ Vip+ cells can be recognized at a relatively lower resolution, Cck+ cells can be further divided into Cck+/ C1ql3+ and Cck+/ Bdnf+ subtypes at a higher resolution. c, Heatmap of marker gene expression in 5 SCN neuron subtypes in Drop-seq data. d, UMAP plot showing the integration of Drop-seq data and 10X data. (e) Composition of SCN neuron subtypes from Drop-seq data in SCN neuron subtypes from 10X dataset after mapping Drop-seq data to 10X data. f, Correlation of marker gene expression profiles between Drop-seq data to 10X data. Two-sided p-value was determined by correlation test. g, Violin plot showing the expression profiles of marker genes of 5 SCN neuron subtypes in 10X dataset. h, Circadian phases of core clock genes in different SCN neuron subtypes. The rectangular bars represent the estimated phase range, i.e. φ±SEM, where SEM stands for Standard Error of Mean. Two-sided p-values of pair-wise comparisons of phase difference are represented as * p< 0.05, ** p<0.01, *** p<0.001. P-values were calculated as described in methods. Two circadian cycles include 12 circadian datapoints were used to calculate p-values.
Extended Data Fig. 5 Analysis of circadian gene expression in SCN neuron subtypes.
a, Circadian rhythmicity of 5 SCN neuron subtypes and 10 non-SCN neuron subtypes. Rhythmicity of core clock genes (left). Percentage of rhythmic genes in total expressed genes in each neuron subtype (right). N14 and N15 subtypes were excluded in this analysis due to their small number of cells. P-values were calculated with JTKCycle. b, smFISH showing the expression patterns of Grp (green) and Bmal1 (red) in SCN at ZT14. DAPI signals in blue indicate cell body positions. Scale bar is 100 μm. c, Quantification and comparison of Bmal1 expression among Grp+ cells within SCN (n=87), Grp- cells within SCN (n=711) and non-SCN cells (n=1521). The boxplots indicate the minimum, maximum, first quartile, third quartile and outliers of the distribution of single-cell Bmal1 expression. p-values are calculated by ANOVA with Dunnett correction. d, smFISH showing the expression of Per2 (red), Vip (green) and Cck (white) in SCN at 6 circadian time points. Scale bar represents 10μm. e, Quantification of circadian expression of Per2 in Vip+ and Cck+ neurons by the number of Per2 counts in each type of neurons in SCN. The boxplots indicate the median, minimum, maximum, first quartile and third quartile of the distribution of single-cell Per2 expression. Circadian phase (φ) and its error were analyzed by cosine regression on the population median values of each subtype at each time point. The numbers of Vip+ neurons used in the analysis: n=178 (CT14); n=197 (CT18); n=240 (CT22); n=195 (CT26); n=229 (CT30); n=206 (CT34). The numbers of Cck+ neurons used in the analysis: n=226 (CT14); n=250 (CT18); n=305 (CT22); n=248 (CT26); n=291 (CT30); n=262 (CT34). P-values of comparisons between peak and trough values using two-sided Mann-Whitney U-test are p=1.3x10-38 (Vip+) and p=7.3x10-10 (Cck+). P-values are represented as * p<0.05, ** p<0.01, *** p<0.001.
Extended Data Fig. 6 Cell-type-specific light-affected gene expression and spatial gene expression in SCN.
a, Comparison of log2-transformed fold changes of 92 overlapped light-affected genes between SCN neurons in this paper and those from Hatori et al.’s bulk SCN data. Light-affected genes in our data and their log2-transformed fold changes used here were obtained by Monocle program. Pearson’s correlation coefficients were indicated in the Figure. b, Pairwise comparison of the differences of log2-transformed fold changes among Avp+/Nms+, Vip+/Nms+ and Grp+/Vip+ subtypes. Cck+/C1ql3+ and Cck+/Bdnf+ subtypes were not included in this analysis because their low response to light stimulation (see text). The boxplots indicate the minimum, median, maximum, first quartile and third quartile. Two-sided p-values were calculated by t-test. There are 29, 21 and 20 genes in Vip+/Nms+ vs. Avp+/Nms+ group, Grp+/Vip+ vs. Vip+/Nms+ group, and Grp+/Vip+ vs. Avp+/Nms+ group, respectively. (c) Percentage of light-affected genes in total expressed genes in all major cell types in SCN (FDR<0.05; upregulated genes in red and down-regulated in green). FDR was calculated by Monocle program. d, Comparison of spatial distribution of gene expression in SCN between LCM RNA-seq data and tissue clearing imaging data. LCM samples and cell bodies in tissue-clearing data were divided into 3 core-shell subdivisions, 6 anterior-posterior subdivisions and 2 medial-lateral subdivisions respectively. The center of SCN in core-shell division is chosen at the half point on medial-lateral axis and ventral quarter point on dorsal-ventral axis of the maximum SCN cross-section on anterior-posterior axis. e, Expression of neuropeptides (gene symbols in blue) and receptors (gene symbols in green) that are specifically expressed in SCN neuron subtypes across circadian time points. The rhythmic genes coding for neuropeptides and receptors were highlighted in red.
Supplementary information
Supplementary Tables.
Supplementary Tables 1–6.
Supplementary video 1
Subtype-specific spatial domains reconstructed in 3D SCN. Vip+ domain (pink), Grp+ domain (cyan), Cck+ domain (purple) and Avp+ domain (yellow).
Supplementary video 2
A guide on how to use the SCN 3D atlas. Users can select specific spatial domains to be visualized in 3D as well as search for the expression patterns of specific genes in LCM-seq data.
Rights and permissions
About this article
Cite this article
Wen, S., Ma, D., Zhao, M. et al. Spatiotemporal single-cell analysis of gene expression in the mouse suprachiasmatic nucleus. Nat Neurosci 23, 456–467 (2020). https://doi.org/10.1038/s41593-020-0586-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-020-0586-x
This article is cited by
-
Neuronal and glial vulnerability of the suprachiasmatic nucleus in tauopathies: evidence from human studies and animal models
Molecular Neurodegeneration (2024)
-
System-level time computation and representation in the suprachiasmatic nucleus revealed by large-scale calcium imaging and machine learning
Cell Research (2024)
-
Maternal dietary fat during lactation shapes single nucleus transcriptomic profile of postnatal offspring hypothalamus in a sexually dimorphic manner in mice
Nature Communications (2024)
-
Single-cell transcriptomics reveals that glial cells integrate homeostatic and circadian processes to drive sleep–wake cycles
Nature Neuroscience (2024)
-
Integrated neural tracing and in-situ barcoded sequencing reveals the logic of SCN efferent circuits in regulating circadian behaviors
Science China Life Sciences (2024)