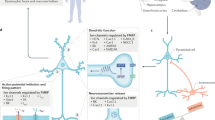
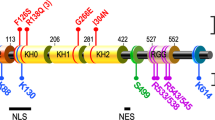
Abstract
Fragile X mental retardation protein (FMRP) is the product of the fragile X mental retardation 1 gene (FMR1), a gene that — when epigenetically inactivated by a triplet nucleotide repeat expansion — causes the neurodevelopmental disorder fragile X syndrome (FXS). FMRP is a widely expressed RNA-binding protein with activity that is essential for proper synaptic plasticity and architecture, aspects of neural function that are known to go awry in FXS. Although the neurophysiology of FXS has been described in remarkable detail, research focusing on the molecular biology of FMRP has only scratched the surface. For more than two decades, FMRP has been well established as a translational repressor; however, recent whole transcriptome and translatome analyses in mouse and human models of FXS have shown that FMRP is involved in the regulation of nearly all aspects of gene expression. The emerging mechanistic details of the mechanisms by which FMRP regulates gene expression may offer ways to design new therapies for FXS.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Nelson, D. L., Orr, H. T. & Warren, S. T. The unstable repeats–three evolving faces of neurologic disease. Neuron 77, 825–843 (2013).
Wang, L. W., Berry-Kravis, E. & Hagerman, R. J. Fragile X: leading the way for targeted treatments in autism. Neurotherapeutics 7, 264–274 (2010).
Pieretti, M. et al. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 66, 817–822 (1991).
Verkerk, A. J. et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65, 905–914 (1991).
Schaefer, G. B. & Mendelsohn, N. J. Genetics evaluation for the etiologic diagnosis of autism spectrum disorders. Genet. Med. 10, 4–12 (2008).
Li, Y. & Zhao, X. Concise review: Fragile X proteins in stem cell maintenance and differentiation. Stem Cell 32, 1724–1733 (2014).
Liu, B. et al. Regulatory discrimination of mRNAs by FMRP controls mouse adult neural stem cell differentiation. Proc Natl Acad Sci USA 115, E11397–E11405 (2018).
Berry-Kravis, E. M. et al. Drug development for neurodevelopmental disorders: lessons learned from fragile X syndrome. Nat. Rev. Drug Discov. 17, 280–299 (2018).
Zhao, X. & Bhattacharyya, A. Human models are needed for studying human neurodevelopmental disorders. Am. J. Hum. Genet. 103, 829–857 (2018).
Ashley, C. T. Jr., Wilkinson, K. D., Reines, D. & Warren, S. T. FMR1 protein: conserved RNP family domains and selective RNA binding. Science 262, 563–566 (1993).
Siomi, H., Siomi, M. C., Nussbaum, R. L. & Dreyfuss, G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell 74, 291–298 (1993).
Darnell, J. C. et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011). This article shows that FMRP ‘CLIPs’ to nearly 1,000 RNAs in the mouse brain, many of which encode proteins involved in synaptic function and are relevant to autism. In addition, it shows that most of the FMRP CLIP sites are in CDS of mRNA.
Udagawa, T. et al. Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat. Med. 19, 1473–1477 (2013).
Richter, J. D., Bassell, G. J. & Klann, E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat. Rev. Neurosci. 16, 595–605 (2015).
Hagerman, R. J. et al. Fragile X syndrome. Nat. Rev. Dis. Prim. 3, 17065 (2017).
Gross, C., Hoffmann, A., Bassell, G. J. & Berry-Kravis, E. M. Therapeutic strategies in fragile X syndrome: from bench to bedside and back. Neurotherapeutics 12, 584–608 (2015).
Bakker, C. E. & Oostra, B. A. Understanding fragile X syndrome: insights from animal models. Cytogenet. Genome Res. 100, 111–123 (2003).
Bhattacharyya, A. & Zhao, X. Human pluripotent stem cell models of fragile X syndrome. Mol. Cell Neurosci. 73, 43–51 (2016).
Garber, K. B., Visootsak, J. & Warren, S. T. Fragile X syndrome. Eur. J. Hum. Genet. 16, 666–672 (2008).
Sansone, S. M. et al. Improving IQ measurement in intellectual disabilities using true deviation from population norms. J. Neurodev. Disord. 6, 16 (2014).
Kaufmann, W. E. Autism spectrum disorder in fragile X syndrome: concurring conditions and current treatment. Pediatrics 139, S194–S206 (2017).
Shah, S. et al. FMRP control of ribosome translocation promotes chromatin modifications and alternative splicing of neuronal genes linked to autism. Cell Rep. 30, 4459–4472 (2020). The authors use dynamic ribosome profiling to identify mRNAs that are associated with FMRP-stalled ribosomes. A number of these RNAs encode chromatin-modifying enzymes, one of which alters histone H3 trimethylation at lysine 36 and alternative splicing of pre-mRNA.
Tan, M. H. et al. Dynamic landscape and regulation of RNA editing in mammals. Nature 550, 249–254 (2017).
Myrick, L. K., Hashimoto, H., Cheng, X. & Warren, S. T. Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain. Hum. Mol. Genet. 24, 1733–1740 (2015).
Adinolfi, S. et al. The N-terminus of the fragile X mental retardation protein contains a novel domain involved in dimerization and RNA binding. Biochemistry 42, 10437–10444 (2003).
Ramos, A. et al. The structure of the N-terminal domain of the fragile X mental retardation protein: a platform for protein–protein interaction. Structure 14, 21–31 (2006).
Eberhart, D. E., Malter, H. E., Feng, Y. & Warren, S. T. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum. Mol. Genet. 5, 1083–1091 (1996).
Ceman, S. et al. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum. Mol. Genet. 12, 3295–3305 (2003).
Huang, J., Ikeuchi, Y., Malumbres, M. & Bonni, A. A Cdh1-APC/FMRP ubiquitin signaling link drives mGluR-dependent synaptic plasticity in the mammalian rain. Neuron 86, 726–739 (2016).
Muddashetty, R. S. et al. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation and mGluR signaling. Mol. Cell 42, 673–688 (2012).
Kim, T. H. et al. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 365, 825–829 (2019).
Tsang, B. et al. Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation. Proc. Natl Acad. Sci. USA 116, 4218–4227 (2019).
Althar, Y. M. & Joseph, S. RNA binding specificity of the human fragile X mental retardation protein. J. Mol. Biol. 432, 3851–3868 (2020).
Darnell, J. C. et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 107, 489–499 (2001).
Vasilyev, N. et al. Crystal structure reveals specific recognition of a G-quadruplex RNA by a β-turn in the RGG motif of FMRP. Proc. Natl Acad. Sci. USA 112, E5391–E5400 (2015).
Darnell, J. C. et al. Kissing complex RNAs mediate interaction between the fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 19, 903–918 (2005).
Darnell, J. C., Fraser, C. E., Mostovetsky, O. & Darnell, R. B. Discrimination of common and unique RNA-binding activities among fragile X mental retardation protein paralogs. Hum. Mol. Genet. 18, 3164–3177 (2009).
Brown, V. et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107, 477–487 (2001).
Miyashiro, K. Y. et al. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron 37, 417–431 (2003).
Maurin, T. et al. HITS-CLIP in various brain areas reveals new targets and new modalities of RNA binding by fragile X mental retardation protein. Nucleic Acids Res. 46, 6344–6355 (2018).
Van Driesche, S. J. et al. FMRP binding to a ranked subset of long genes is revealed by coupled CLIP and TRAP in specific neuronal cell types. Preprint at bioRxiv https://doi.org/10.1101/762500 (2019).
Sawicka, K. et al. FMRP has a cell-type-specific role in CA1 pyramidal neurons to regulate autism-related transcripts and circadian memory. eLife 8, e46919 (2019). The authors use FMRP conditional tag mice to perform cell type-specific FMRP target identification in mouse CA1 and cerebellar granule neurons and discover novel cell type-specific targets of FMRP.
Zhang, J. et al. Fragile X-related proteins regulate mammalian circadian behavioral rhythms. Am. J. Hum. Genet. 83, 43–52 (2008).
Kwan, K. Y. et al. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell 149, 899–911 (2012).
Doers, M. E. et al. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cell Dev. 23, 1777–1787 (2014).
Khalfallah, O. et al. Depletion of the fragile X mental retardation protein in embryonic stem cells alters the kinetics of neurogenesis. Stem Cell 35, 374–385 (2017).
Telias, M., Mayshar, Y., Amit, A. & Ben-Yosef, D. Molecular mechanisms regulating impaired neurogenesis of fragile X syndrome human embryonic stem cells. Stem Cell Dev. 24, 2353–2365 (2015).
Ascano, M. Jr. et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492, 382–386 (2012).
Tran, S. S. et al. Widespread RNA editing dysregulation in brains from autistic individuals. Nat. Neurosci. 22, 25–36 (2019). This article demonstrates that adenosine-to-inosine RNA editing is reduced in ASD and FXS brains and that FMRP modulates RNA editing through interaction with the RNA-editing enzyme ADAR. It also identifies FMRP target RNAs in adult human brains by CLIP–seq.
Li, M. et al. Identification of FMR1-regulated molecular networks in human neurodevelopment. Genome Res. 30, 361–374 (2020). The authors identify FMRP-binding targets using CLIP–seq in pure populations of human NPCs and neurons in either excitatory or inhibitory lineages differentiated from human pluripotent stem cells. They show both shared and unique FMRP targets in different cell types and human-specific FMRP targets.
Chasman, D., Fotuhi Siahpirani, A. & Roy, S. Network-based approaches for analysis of complex biological systems. Curr. Opin. Biotechnol. 39, 157–166 (2016).
Anderson, B. R., Chopra, P., Suhl, J. A., Warren, S. T. & Bassell, G. J. Identification of consensus binding sites clarifies FMRP binding determinants. Nucleic Acids Res. 44, 6649–6659 (2016).
Suhl, J. A., Chopra, P., Anderson, B. R., Bassell, G. J. & Warren, S. T. Analysis of FMRP mRNA target datasets reveals highly associated mRNAs mediated by G-quadruplex structures formed via clustered WGGA sequences. Hum. Mol. Genet. 23, 5479–5491 (2014).
King, I. F. et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature 501, 58–62 (2013).
Ahmad, M. et al. Topoisomerase 3beta is the major topoisomerase for mRNAs and linked to neurodevelopment and mental dysfunction. Nucleic Acids Res. 45, 2704–2713 (2017).
Lee, S. K. et al. Topoisomerase 3beta interacts with RNAi machinery to promote heterochromatin formation and transcriptional silencing in Drosophila. Nat. Commun. 9, 4946 (2018).
Xu, D. et al. Top3beta is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci. 16, 1238–1247 (2013).
Feng, Y. et al. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell 1, 109–118 (1997).
Khandjian, E. W., Corbin, F., Woerly, S. & Rousseau, F. The fragile X mental retardation protein is associated with ribosomes. Nat. Genet. 12, 91–93 (1996).
Dölen, G. et al. Correction of fragile X syndrome in mice. Neuron 56, 955–962 (2007).
Thomson, S. R. et al. Cell-type-specific translation profiling reveals novel strategy for treating fragile X syndrome. Neuron 95, 550–563 (2017). This work uses TRAP–seq to identify cell type-specific translational changes in hippocampal CA1 neurons as a result of FMRP deficiency. The authors identify muscarinic acetylcholine receptor 4 as a novel drug target to alleviate FMRP deficiency.
Ceolin, L. et al. Cell type-specific mRNA dysregulation in hippocampal CA1 pyramidal neurons of the fragile X syndrome mouse model. Front. Mol. Neurosci. 10, 340 (2017).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2012).
Liu, B. et al. Optimization of ribosome profiling using low-input brain tissue from fragile X syndrome model mice. Nucleic Acids Res. 47, e25 (2019).
Das Sharma, S. et al. Widespread alterations in translation elongation in the brain of juvenile Fmr1 knockout mice. Cell Rep. 26, 3313–3322 (2019).
Greenblatt, E. J. & Spradling, A. C. Fragile X mental retardation 1 gene enhances the translation of large autism-related proteins. Science 361, 709–712 (2018).
Shu, S. et al. FMRP links optimal codons to mRNA stability in neurons. Proc. Natl Acad. Sci. USA 117, 30400–30411 (2020). This work uses ribosome profiling and metabolic labelling of RNA to demonstrate that FMRP influences the relationship between codon optimality and RNA stability.
Hien, A., Molinaro, G., Liu, B., Huber, K. M. & Richter, J. D. Ribosome profiling in mouse hippocampus: plasticity-induced regulation and bidirecitonal controlby TSC2 and FMRP. Mol. Autism 14, 78 (2020).
Luo, Y. et al. Fragile X mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet. 6, e1000898 (2010).
Guo, W. et al. Inhibition of GSK3beta improves hippocampus-dependent learning and rescues neurogenesis in a mouse model of fragile X syndrome. Hum. Mol. Genet. 21, 681–691 (2012).
El-Brolosy, M. A. & Stainier, D. Y. R. Genetic compensation: a phenomenon in search of mechanisms. PLoS Genet. 13, e1006780 (2017).
Liu, T. et al. Direct measurement of the mechanical work during translocation by the ribosome. eLife 3, e03406 (2014).
Richter, J. D. & Coller, J. Pausing on polysomes: make way for elongation in translational control. Cell 16, 292–300 (2015).
Hanson, G. & Coller, J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell. Biol. 19, 20–30 (2018).
Radhakrishnan, A. & Green, R. Connections underlying translation and mRNA stability. J. Mol. Biol. 428, 3558–3564 (2016).
Heck, A. M. & Wilusz, J. The interplay between the RNA decay and translation machinery in eukaryotes. Cold Spring Harb. Perspect. Biol. 10, a032839 (2018).
Zalfa, F. et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat. Neurosci. 10, 578–587 (2007).
Zhang, F. et al. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 27, 3936–3950 (2018).
Didiot, M. C., Subramanian, M., Flatter, E., Mandel, J. L. & Moine, H. Cells lacking the fragile X mental retardation protein (FMRP) have normal RISC activity but exhibit altered stress granule assembly. Mol. Biol. Cell 20, 428–437 (2009).
Valdez-Sinon, A. N. et al. Cdh1-APC regulates protein synthesis and stress granules in neurons through an FMRP-dependent mechanism. iScience 23, 101132 (2020).
Stoecklin, G. & Kedersha, N. Relationship of GW/P-bodies with stress granules. Adv. Exp. Med. Biol. 768, 197–211 (2013).
Perez-Ortin, J. E., Alepus, P., Chavez, S. & Choder, M. Eukaryotic mRNA decay: methodologies, pathways, and links to other stages of gene expression. J. Mol. Biol. 425, 3750–3775 (2013).
Jin, P. et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 7, 113–117 (2004).
Kenny, P. J. et al. MOV10 and FMRP regulate AGO2 association with microRNA recognition elements. Cell Rep. 9, 1729–1741 (2014).
Chen, E., Sharma, M. R., Shi, X., Agrawal, R. K. & Joseph, S. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol. Cell 54, 407–417 (2014).
Schenck, A., Bardoni, B., Moro, A., Bagni, C. & Mandel, J. L. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc. Natl Acad. Sci. USA 98, 8844–8849 (2001).
De Rubeis, S. et al. CYFIP1 Coordinates mRNA translation and cytoskeleton remodeling to ensure proper dendritic spine formation. Neuron 79, 1169–1182 (2013).
Abekhoukh, S. et al. New insights into the regulatory function of CYFIP1 in the context of WAVE- and FMRP-containing complexes. Dis. Model. Mech. 10, 463–474 (2017).
Meyer, K. D. & Jaffrey, S. R. Rethinking m6A readers, writers, and erasers. Ann. Rev. Cell Dev. Biol. 33, 319–342 (2017).
Zhao, B. S., Roundtree, I. A. & He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42 (2017).
Edupuganti, R. R. et al. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878 (2017).
Edens, B. M. et al. FMRP modulates neural differentiation through m6A-dependent mRNA nuclear export. Cell Rep. 28, 845–854 (2019). This article shows that FMRP preferentially binds m6A-methylated mRNAs and facilitates their nuclear export, which is important for NPC differentiation into neurons.
Engel, M. et al. The role of m6A/m-RNA methylation in stress response regulation. Neuron 99, 389–403 (2018).
Reich, D. P. & Bass, B. L. Mapping the dsRNA world. Cold Spring Harb. Perspect. Biol. 11, a035352 (2019).
Bhogal, B. et al. Modulation of dADAR-dependent RNA editing by the Drosophila fragile X mental retardation protein. Nat. Neurosci. 14, 1517–1524 (2011).
Shamay-Ramot, A. et al. Fmrp interacts with Adar and regulates RNA editing, synaptic density and locomotor activity in zebrafish. PLoS Genet. 11, e1005702 (2015).
McMahon, A. C. et al. TRIBE: hijacking an RNA-editing enzyme to identify cell-specific targets of RNA-binding proteins. Cell 165, 742–753 (2016).
Korb, E. et al. Excess translation of epigenetic regulators contributes to fragile X syndrome and is alleviated by Brd4 inhibition. Cell 170, 1209–1223 (2017). This article demonstrates that FMRP controls the synthesis of epigenetic modifying enzymes that in turn alter the chromatin landscape, leading to transcriptional activation. The authors also use a small-molecule inhibitor of one these proteins and demonstrate rescue of several FXS-related phenotypes in mice.
Li, Y. et al. Reducing histone acetylation rescues cognitive deficits in a mouse model of Fragile X syndrome. Nat. Commun. 9, 2494 (2018).
Alpatov, R. et al. A chromatin-dependent role of the fragile X mental retardation protein FMRP in the DNA damage response. Cell 157, 869–881 (2014).
Guo, J. U. & Bartel, D. P. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science 353, aaf5371 (2016).
Pasciuto, E. & Bagni, C. SnapShot: FMRP interacting proteins. Cell 159, 218 (2014).
Lombard-Banek, C., Choi, S. B. & Nemes, P. Single-cell proteomics in complex tissue using microprobe capillary electrophoresis mass spectrometry. Methods Enzymol. 628, 263–292 (2019).
Schafer, S. T. et al. Pathological priming causes developmental gene network heterochronicity in autistic subject-derived neurons. Nat. Neurosci. 22, 243–255 (2019).
Protic, D., Salcedo-Arellanom, M. J., Dym, J. B., Potter, L. A. & Hagerman, R. J. New targeted treatments for fragile X syndrome. Curr. Pediatr. Rev. 15, 251–258 (2019).
Gantois, I., Popic, J., Khoutorsky, A. & Sonenberg, N. Metformin for treatment of fragile X syndrome and other neurological disorders. Ann. Rev. Med. 70, 167–181 (2019).
Brown, V. et al. Purified recombinant Fmrp exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein. J. Biol. Chem. 273, 15521–15527 (1998).
Devys, D., Lutz, Y., Rouyer, N., Bellocq, J. P. & Mandel, J. L. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat. Genet. 4, 335–340 (1993).
Heiman, M. et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748 (2008).
Men, Y. et al. Astroglial FMRP deficiency cell-autonomously up-regulates miR-128 and disrupts developmental astroglial mGluR5 signaling. Proc. Natl Acad. Sci. USA 117, 25092–25103 (2020).
Jacobs, S., Cheng, C. & Doering, L. C. Hippocampal neuronal subtypes develop abnormal dendritic arbors in the presence of fragile X astrocytes. Neuroscience 324, 202–217 (2016).
Nyquist, P. A. & Hagerman, R. Genetics, white matter, and cognition: the effects of methylation on FMR1. Neurology 88, 2070–2071 (2017).
Wang, H. et al. Developmentally-programmed FMRP expression in oligodendrocytes: a potential role of FMRP in regulating translation in oligodendroglia progenitors. Hum. Mol. Genet. 13, 79–89 (2004).
Acknowledgements
Work in the authors’ laboratories was supported by the NIH (U54HD082013, GM046779 and GM135087 to J.D.R., R01MH116582, MH118827 and R01NS105200 to X.Z and U54HD090256 to the Waisman Center), the Simons Foundation (to J.D.R.) and a Jenni and Kyle Professorship to X.Z.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Neuroscience thanks B. Bardoni; R. Kelleher; E. Schuman; and N. Sonenberg, who co-reviewed with I. Gantois, for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- RNA-binding protein
-
Member of a family of proteins that frequently contain certain amino acid sequence motifs that have a strong avidity for single-stranded or double-stranded RNA.
- Human pluripotent stem cells
-
Cells that have the capability to differentiate into any cell of the body.
- Gene editing
-
Alteration (by insertion, deletion or substitution) of the nucleotide sequence within a gene, frequently by way of CRISPR–Cas9 technology.
- mRNA splicing
-
The process by which precursor RNA is processed into mature RNA through removal of intron-derived sequences.
- Next-generation sequencing
-
Massively parallel and ultra-high-throughput sequencing.
- Gene network analysis
-
Analysis of gene involvement in two or more processes, which in turn may contain many genes that contribute to a given process.
- mRNA codon bias
-
A situation where an mRNA does not contain all codons for a given amino acid in equal proportions.
- N 6-Methyladenosine
-
(m6A). Adenosine methylated at the nitrogen at the 6 position.
- Phase separation
-
Physical state of a protein that can form a membraneless separation between liquid and gel-like phases.
- Secondary structures
-
In RNA, intramolecular base pairing.
- Network-based integrative analysis
-
A process that seeks to integrate differential gene expression patterns based on calculated probability values with network-based meta-analysis to identify patterns of genes and pathways that may be impacted by underlying biological conditions such as disease.
- Topoisomerase
-
An enzyme that unwinds DNA.
- Ribosome translocation
-
The movement of a ribosome in the 5′ to 3′ direction as it translates mRNA sequence information into a polypeptide.
- Stress granule
-
A membraneless subcellular organelle that forms in response to cellular stress and contains a variety of RNAs and proteins.
- DNA damage response
-
The response of cells to identify and repair broken DNA strands.
Rights and permissions
About this article
Cite this article
Richter, J.D., Zhao, X. The molecular biology of FMRP: new insights into fragile X syndrome. Nat Rev Neurosci 22, 209–222 (2021). https://doi.org/10.1038/s41583-021-00432-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41583-021-00432-0
This article is cited by
-
FMRP-mediated spatial regulation of physiologic NMD targets in neuronal cells
Genome Biology (2024)
-
RNA modifications in physiology and disease: towards clinical applications
Nature Reviews Genetics (2024)
-
PGC-1α integrates insulin signaling with mitochondrial physiology and behavior in a Drosophila model of Fragile X Syndrome
npj Metabolic Health and Disease (2024)
-
The multifaceted role of Fragile X-Related Protein 1 (FXR1) in cellular processes: an updated review on cancer and clinical applications
Cell Death & Disease (2024)
-
Differential cognitive and behavioral development from 6 to 24 months in autism and fragile X syndrome
Journal of Neurodevelopmental Disorders (2024)