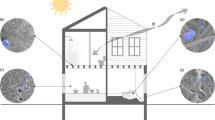
Abstract
The built environment comprises all structures built by humans, including our homes, workplaces, schools and vehicles. As in any ecosystem on Earth, microorganisms have been found in every part of the built environment that has been studied. They exist in the air, on surfaces and on building materials, usually dispersed by humans, animals and outdoor sources. Those microbial communities and their metabolites have been implied to cause (or exacerbate) and prevent (or mitigate) human disease. In this Review, we outline the history of the field of microbiology of the built environment and discuss recent insights that have been gained into microbial ecology, adaptation and evolution of this ecosystem. Finally, we consider the implications of this research, specifically, how it is changing the types of materials we use in buildings and how our built environments affect human health.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Martin, L. J. et al. Evolution of the indoor biome. Trends Ecol. Evol. 30, 223–232 (2015).
Klepeis, N. E. et al. The National Human Activity Pattern Survey (NHAPS): a resource for assessing exposure to environmental pollutants. J. Expo. Anal. Environ. Epidemiol. 11, 231–252 (2001).
Gibbons, S. M. The built environment is a microbial wasteland. mSystems 1, e00033 (2016).
Kelley, S. T., Theisen, U., Angenent, L. T., St Amand, A. & Pace, N. R. Molecular analysis of shower curtain biofilm microbes. Appl. Environ. Microbiol. 70, 4187–4192 (2004). This is the first study to apply molecular amplicon sequencing approaches to the actual microbiome of the built environment.
Gibbons, S. M. et al. Ecological succession and viability of human-associated microbiota on restroom surfaces. Appl. Environ. Microbiol. 81, 765–773 (2015).
Kembel, S. W. et al. Architectural design influences the diversity and structure of the built environment microbiome. ISME J. 6, 1469–1479 (2012).
Adams, R. I., Miletto, M., Taylor, J. W. & Bruns, T. D. Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 7, 1262–1273 (2013).
Flores, G. E. et al. Diversity, distribution and sources of bacteria in residential kitchens: bacterial diversity of residential kitchens. Environ. Microbiol. 15, 588–596 (2013).
Dunn, R. R., Fierer, N., Henley, J. B., Leff, J. W. & Menninger, H. L. Home life: factors structuring the bacterial diversity found within and between homes. PLOS ONE 8, e64133 (2013).
Bokulich, N. A., Ohta, M., Richardson, P. M. & Mills, D. A. Monitoring seasonal changes in winery-resident microbiota. PLOS ONE 8, e66437 (2013).
Jeon, Y.-S., Chun, J. & Kim, B.-S. Identification of household bacterial community and analysis of species shared with human microbiome. Curr. Microbiol. 67, 557–563 (2013).
Lax, S. et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048–1052.This study applies longitudinal analysis of both the human and animal occupants and built surfaces in homes, providing the first examples of the intensity of bidirectional colonization events.
Miletto, M. & Lindow, S. E. Relative and contextual contribution of different sources to the composition and abundance of indoor air bacteria in residences. Microbiome 3, 61 (2015).
Chase, J. et al. Geography and location are the primary drivers of office microbiome composition. mSystems 1, e00022-16 (2016). This study shows that under normal (dry) office conditions, bacterial communities do not differ on the basis of material but do differ on the basis of the location in a room.
Adams, R. I., Bateman, A. C., Bik, H. M. & Meadow, J. F. Microbiota of the indoor environment: a meta-analysis. Microbiome 3, 49 (2015).
Lee, L., Tin, S. & Kelley, S. T. Culture-independent analysis of bacterial diversity in a child-care facility. BMC Microbiol. 7, 27 (2007).
Rintala, H., Pitkaranta, M., Toivola, M., Paulin, L. & Nevalainen, A. Diversity and seasonal dynamics of bacterial community in indoor environment. BMC Microbiol. 8, 56 (2008).
Hewitt, K. M., Gerba, C. P., Maxwell, S. L. & Kelley, S. T. Office space bacterial abundance and diversity in three metropolitan areas. PLOS ONE 7, e37849 (2012).
Hewitt, K. M. et al. Bacterial diversity in two neonatal intensive care units (NICUs). PLOS ONE 8, e54703 (2013).
Kembel, S. W. et al. Architectural design drives the biogeography of indoor bacterial communities. PLOS ONE 9, e87093 (2014).
Meadow, J. F. et al. Bacterial communities on classroom surfaces vary with human contact. Microbiome 2, 7 (2014).
Meadow, J. F. et al. Humans differ in their personal microbial cloud. PeerJ. 3, e1258 (2015).
Amend, A. S., Seifert, K. A., Samson, R. & Bruns, T. D. Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proc. Natl Acad. Sci. USA 107, 13748–13753 (2010). This is the first study to explore the geographical and climate-driven diversity of indoor fungal communities, providing evidence to support the potential relationship between fungal diversity and composition and differences in health characteristics.
Barberán, A. et al. The ecology of microscopic life in household dust. Proc. R. Soc. B Biol. Sci. 282, 20151139 (2015).
Emerson, J. B. et al. Impacts of flood damage on airborne bacteria and fungi in homes after the 2013 Colorado Front Range flood. Environ. Sci. Technol. 49, 2675–2684 (2015).
Flores, G. E. et al. Microbial biogeography of public restroom surfaces. PLOS ONE 6, e28132 (2011).
Dannemiller, K. C., Gent, J. F., Leaderer, B. P. & Peccia, J. Influence of housing characteristics on bacterial and fungal communities in homes of asthmatic children. Indoor Air 26, 179–192 (2016).
Weikl, F. et al. Fungal and bacterial communities in indoor dust follow different environmental determinants. PLOS ONE 11, e0154131 (2016).
Emerson, J. B. et al. High temporal variability in airborne bacterial diversity and abundance inside single-family residences. Indoor Air 27, 576–586 (2017).
Qian, J., Hospodsky, D., Yamamoto, N., Nazaroff, W. W. & Peccia, J. Size-resolved emission rates of airborne bacteria and fungi in an occupied classroom: size-resolved bioaerosol emission rates. Indoor Air 22, 339–351 (2012). This study involves a fundamental quantitative analysis of the abundance of bacterial and fungal particles emitted by occupants in a built environment, which supports the important contribution of humans and animals to the discovered diversity of indoor microbial communities.
Hospodsky, D. et al. Human occupancy as a source of indoor airborne bacteria. PLOS ONE 7, e34867 (2012).
Hospodsky, D. et al. Characterizing airborne fungal and bacterial concentrations and emission rates in six occupied children’s classrooms. Indoor Air 25, 641–652 (2015).
Kunkel, S. A., Azimi, P., Zhao, H., Stark, B. C. & Stephens, B. Quantifying the size-resolved dynamics of indoor bioaerosol transport and control. Indoor Air 27, 977–987 (2017).
Miller, J. D. & Young, J. C. The use of ergosterol to measure exposure to fungal propagules in indoor air. Am. Ind. Hyg. Assoc. J. 58, 39–43 (1997).
Rao, C. Y., Cox-Ganser, J. M., Chew, G. L., Doekes, G. & White, S. Use of surrogate markers of biological agents in air and settled dust samples to evaluate a water-damaged hospital. Indoor Air 15, 89–97 (2005).
Vandeputte, D. et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551, 507–511 (2017).
Dannemiller, K. C., Lang-Yona, N., Yamamoto, N., Rudich, Y. & Peccia, J. Combining real-time PCR and next-generation DNA sequencing to provide quantitative comparisons of fungal aerosol populations. Atmos. Environ. 84, 113–121 (2014).
Props, R. et al. Absolute quantification of microbial taxon abundances. ISME J. 11, 584–587 (2017).
Prussin, A. J. & Marr, L. C. Sources of airborne microorganisms in the built environment. Microbiome 3, 78 (2015).
Prussin, A. J., Garcia, E. B. & Marr, L. C. Total concentrations of virus and bacteria in indoor and outdoor air. Environ. Sci. Technol. Lett. 2, 84–88 (2015).
Tringe, S. G. et al. The airborne metagenome in an indoor urban environment. PLOS ONE 3, e1862 (2008). This is the first metagenomics study of the indoor microbiome that found that the indoor air microorganisms were not random transients from surrounding outdoor environments, but rather originated from indoor niches.
Yooseph, S. et al. A metagenomic framework for the study of airborne microbial communities. PLOS ONE 8, e81862 (2013).
Adams, R. I. et al. Microbes and associated soluble and volatile chemicals on periodically wet household surfaces. Microbiome 5, 128 (2017). This is the first example of the integration of metabolomics and microbiome data in an indoor environment, enabling a clearer understanding of how the household conditions shape microbial metabolism.
Blazewicz, S. J., Barnard, R. L., Daly, R. A. & Firestone, M. K. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 7, 2061–2068 (2013).
Emerson, J. B. et al. Schrödinger’s microbes: tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 5, 86 (2017).
Miller, J. D. & McMullin, D. R. Fungal secondary metabolites as harmful indoor air contaminants: 10 years on. Appl. Microbiol. Biotechnol. 98, 9953–9966 (2014). This study is a synthesis of available data on the low-molecular-weight toxins reliably known from fungi common on damp building materials and the toxins that have been measured on mouldy building materials.
Hegarty, B., Dannemiller, K. & Peccia, J. Gene expression of indoor fungal communities under damp building conditions: implications for human health. Indoor Air 28, 548–558 (2018).This is the first example of the application of metatranscriptomics to a built environment system, providing knowledge of how microbial transcription of genes is influenced by damp conditions.
Weis, C. P. et al. Secondary aerosolization of viable Bacillus anthracis spores in a contaminated US Senate Office. JAMA 288, 2853–2858 (2002).
Blatny, J. M. et al. Tracking airborne Legionella and Legionella pneumophila at a biological treatment plant. Environ. Sci. Technol. 42, 7360–7367 (2008).
Nazaroff, W. W., Nicas, M. & Miller, S. L. Framework for evaluating measures to control nosocomial tuberculosis transmission. Indoor Air 8, 205–218 (1998).
Nosanchuk, J. D. et al. Evidence of zoonotic transmission of Cryptococcus neoformans from a pet cockatoo to an immunocompromised patient. Ann. Intern. Med. 132, 205 (2000).
Furcolow, M. L., Menges, R. W. & Larsh, H. W. An epidemic of histoplasmosis involving man and animals. Ann. Intern. Med. 43, 173–181 (1955).
Anderson, K. et al. Aspergillosis in immunocompromised paediatric patients: associations with building hygiene, design, and indoor air. Thorax 51, 256–261 (1996).
Dick, E. C., Jennings, L. C., Mink, K. A., Wartgow, C. D. & Inborn, S. L. Aerosol transmission of rhinovirus colds. J. Infect. Dis. 156, 442–448 (1987).
Myatt, T. A. et al. Detection of airborne rhinovirus and its relation to outdoor air supply in office environments. Am. J. Respir. Crit. Care Med. 169, 1187–1190 (2004).
Fabian, P., Brain, J., Houseman, E. A., Gern, J. & Milton, D. K. Origin of exhaled breath particles from healthy and human rhinovirus-infected subjects. J. Aerosol Med. Pulm. Drug Deliv. 24, 137–147 (2011).
Jones, R. M. & Adida, E. Influenza infection risk and predominate exposure route: uncertainty analysis. Risk Anal. 31, 1622–1631 (2011).
Cowling, B. J. et al. Aerosol transmission is an important mode of influenza A virus spread. Nat. Commun. 4, 1935 (2013).
Lindsley, W. G. et al. Viable influenza A virus in airborne particles from human coughs. J. Occup. Environ. Hyg. 12, 107–113 (2015).
Weber, D. J., Rutala, W. A., Miller, M. B., Huslage, K. & Sickbert-Bennett, E. Role of hospital surfaces in the transmission of emerging health care-associated pathogens: norovirus. Clostridium difficile, and Acinetobacter species. Am. J. Infect. Control 38, S25–S33 (2010).
Otter, J. A., Yezli, S. & French, G. L. The role played by contaminated surfaces in the transmission of nosocomial pathogens. Infect. Control Hosp. Epidemiol. 32, 687–699 (2011).
Lopman, B. et al. Environmental transmission of norovirus gastroenteritis. Curr. Opin. Virol. 2, 96–102 (2012).
Brankston, G., Gitterman, L., Hirji, Z., Lemieux, C. & Gardam, M. Transmission of influenza A in human beings. Lancet Infect. Dis. 7, 257–265 (2007).
Wilkins, D., Leung, M. H. & Lee, P. K. Indoor air bacterial communities in Hong Kong households assemble independently of occupant skin microbiomes: household air bacteria differ from occupant skin. Environ. Microbiol. 18, 1754–1763 (2016).
Meadow, J. F. et al. Indoor airborne bacterial communities are influenced by ventilation, occupancy, and outdoor air source. Indoor Air 24, 41–48 (2014).
Leung, M. H. Y., Wilkins, D., Li, E. K. T., Kong, F. K. F. & Lee, P. K. H. Indoor-air microbiome in an urban subway network: diversity and dynamics. Appl. Environ. Microbiol. 80, 6760–6770 (2014).
Lax, S. et al. Bacterial colonization and succession in a newly opened hospital. Sci. Transl Med. 9, eaah6500 (2017).
Peccia, J. & Kwan, S. E. Buildings, beneficial microbes, and health. Trends Microbiol. 24, 595–597 (2016).
USEPA. Exposure Factors Handbook (US Environmental Protection Agency, 2011).
Eder, W. & von Mutius, E. Hygiene hypothesis and endotoxin: what is the evidence? Curr. Opin. Allergy Clin. Immunol. 4, 113–117 (2004).
Ege, M. J. et al. Exposure to environmental microorganisms and childhood asthma. N. Engl. J. Med. 364, 701–709 (2011).
Stein, M. M. et al. Innate immunity and asthma risk in Amish and Hutterite farm children. N. Engl. J. Med. 375, 411–421 (2016).
O’Connor, G. T. et al. Early-life home environment and risk of asthma among inner-city children. J. Allergy Clin. Immunol. 141, 1468–1475 (2017).
Lynch, S. V. et al. Effects of early-life exposure to allergens and bacteria on recurrent wheeze and atopy in urban children. J. Allergy Clin. Immunol. 134, 593–601 (2014). This study demonstrates the association between indoor microbial allergens and asthmatic diseases, providing a solid association between disease and indoor microbiome.
Fujimura, K. E. et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc. Natl Acad. Sci. USA 111, 805–810 (2014).
Kanchongkittiphon, W., Mendell, M. J., Gaffin, J. M., Wang, G. & Phipatanakul, W. Indoor environmental exposures and exacerbation of asthma: an update to the 2000 Review by the Institute of Medicine. Environ. Health Perspect. 123, 6–20 (2014).
Mendell, M. J., Mirer, A. G., Cheung, K., Tong, M. & Douwes, J. Respiratory and allergic health effects of dampness, mold, and dampness-related agents: a review of the epidemiologic evidence. Environ. Health Perspect. 119, 748–756 (2011). This is a systematic review of the epidemiological evidence for associations between dampness and mould and respiratory and allergic effects in humans. Evident dampness or mould has consistent positive associations with multiple allergic and respiratory effects, but measured microbiological agents in dust have limited suggestive associations.
Fisk, W. J., Lei-Gomez, Q. & Mendell, M. J. Meta-analyses of the associations of respiratory health effects with dampness and mold in homes. Indoor Air 17, 284–296 (2007).
Bush, R. K., Portnoy, J. M., Saxon, A., Terr, A. I. & Wood, R. A. The medical effects of mold exposure. J. Allergy Clin. Immunol. 117, 326–333 (2006).
Portnoy, J. M., Kwak, K., Dowling, P., VanOsdol, T. & Barnes, C. Health effects of indoor fungi. Ann. Allergy. Asthma. Immunol. 94, 313–320 (2005).
Wessén, B. & Schoeps, K. O. Microbial volatile organic compounds—what substances can be found in sick buildings? Analyst 121, 1203–1205 (1996).
Korpi, A., Pasanen, A. L. & Pasanen, P. Volatile compounds originating from mixed microbial cultures on building materials under various humidity conditions. Appl. Environ. Microbiol. 64, 2914–2919 (1998).
Araki, A. et al. Diffusive sampling and measurement of microbial volatile organic compounds in indoor air. Indoor Air 19, 421–432 (2009).
Korpi, A., Pasanen, A.-L., Pasanen, P. & Kalliokoski, P. Microbial growth and metabolism in house dust. Int. Biodeterior. Biodegrad. 40, 19–27 (1997).
Kirjavainen, P. V. et al. Microbial secondary metabolites in homes in association with moisture damage and asthma. Indoor Air 26, 448–456 (2016).
Ezeonu, I. M., Price, D. L., Simmons, R. B., Crow, S. A. & Ahearn, D. G. Fungal production of volatiles during growth on fiberglass. Appl. Environ. Microbiol. 60, 4172–4173 (1994).
Schleibinger, H., Laussmann, D., Bornehag, C.-G., Eis, D. & Rueden, H. Microbial volatile organic compounds in the air of moldy and mold-free indoor environments. Indoor Air 18, 113–124 (2008).
Kuske, M., Romain, A.-C. & Nicolas, J. Microbial volatile organic compounds as indicators of fungi. Can an electronic nose detect fungi in indoor environments? Build. Environ. 40, 824–831 (2005).
Ryan, T. J. & Beaucham, C. Dominant microbial volatile organic compounds in 23 US homes. Chemosphere 90, 977–985 (2013).
Wargo, M. J. & Hogan, D. A. Fungal — bacterial interactions: a mixed bag of mingling microbes. Curr. Opin. Microbiol. 9, 359–364 (2006).
Gilbert, J. A. How do we make indoor environments and healthcare settings healthier? Microb. Biotechnol. 10, 11–13 (2017).
Cruz, M. R., Graham, C. E., Gagliano, B. C., Lorenz, M. C. & Garsin, D. A. Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect. Immun. 81, 189–200 (2013).
Graham, C. E., Cruz, M. R., Garsin, D. A. & Lorenz, M. C. Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc. Natl Acad. Sci. USA 114, 4507–4512 (2017).
Kelley, S. T. & Gilbert, J. A. Studying the microbiology of the indoor environment. Genome Biol. 14, 202 (2013).
Verdier, T., Coutand, M., Bertron, A. & Roques, C. A review of indoor microbial growth across building materials and sampling and analysis methods. Build. Environ. 80, 136–149 (2014).
Dedesko, S. & Siegel, J. A. Moisture parameters and fungal communities associated with gypsum drywall in buildings. Microbiome 3, 71 (2015). This is a critical review of measurable moisture parameters on one of the most common building materials and associations with fungal growth.
Gravesen, S., Nielsen, P. A., Iversen, R. & Nielsen, K. F. Microfungal contamination of damp buildings—examples of risk constructions and risk materials. Environ. Health Perspect. 107(Suppl. 3), 505–508 (1999).
Hoang, C. P., Kinney, K. A., Corsi, R. L. & Szaniszlo, P. J. Resistance of green building materials to fungal growth. Int. Biodeterior. Biodegrad. 64, 104–113 (2010).
Gutarowska, B. Metabolic activity of moulds as a factor of building materials biodegradation. Pol. J. Microbiol. 59, 119–124 (2010).
Good, B. H., McDonald, M. J., Barrick, J. E., Lenski, R. E. & Desai, M. M. The dynamics of molecular evolution over 60,000 generations. Nature 551, 45–50 (2017).
Anesti, V. et al. Molecular detection and isolation of facultatively methylotrophic bacteria, including Methylobacterium podarium sp. nov., from the human foot microflora. Environ. Microbiol. 6, 820–830 (2004).
Frank, D. N. et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104, 13780–13785 (2007).
Zhang, L. et al. Microbiological pattern of arterial catheters in the intensive care unit. BMC Microbiol. 10, 266 (2010).
Sangwan, N. et al. Reconstructing an ancestral genotype of two hexachlorocyclohexane-degrading Sphingobium species using metagenomic sequence data. ISME J. 8, 398–408 (2014).
Hammond, T. G. et al. Effects of microgravity on the virulence of Listeria monocytogenes, Enterococcus faecalis, Candida albicans, and methicillin-resistant Staphylococcus aureus. Astrobiology 13, 1081–1090 (2013).
Wilson, J. W. et al. Space flight alters bacterial gene expression and virulence and reveals a role for global regulator Hfq. Proc. Natl Acad. Sci. USA 104, 16299–16304 (2007).
Allen, C. A., Niesel, D. W. & Torres, A. G. The effects of low-shear stress on adherent-invasive Escherichia coli. Environ. Microbiol. 10, 1512–1525 (2008).
La Duc, M. T. et al. Isolation and characterization of bacteria capable of tolerating the extreme conditions of clean room environments. Appl. Environ. Microbiol. 73, 2600–2611 (2007).
Hampton-Marcell, J. T., Lopez, J. V. & Gilbert, J. A. The human microbiome: an emerging tool in forensics. Microb. Biotechnol. 10, 228–230 (2017).
Lax, S. et al. Forensic analysis of the microbiome of phones and shoes. Microbiome 3, 21 (2015).
Brown, G. Z., Kline, J., Mhuireach, G., Northcutt, D. & Stenson, J. Making microbiology of the built environment relevant to design. Microbiome 4, 6 (2016).
Beans, C. The microbiome of green design. BioScience 66, 801–806 (2016).
Zhang, D., Xu, C., Manwani, D. & Frenette, P. S. Neutrophils, platelets, and inflammatory pathways at the nexus of sickle cell disease pathophysiology. Blood 127, 801–809 (2016).
Gilbert, J. A. et al. Current understanding of the human microbiome. Nat. Med. 24, 392–400 (2018).
Leung, M. H. Y. & Lee, P. K. H. The roles of the outdoors and occupants in contributing to a potential pan-microbiome of the built environment: a review. Microbiome 4, 21 (2016).
Coombs, K., Vesper, S., Green, B. J., Yermakov, M. & Reponen, T. Fungal microbiomes associated with green and non-green building materials. Int. Biodeterior. Biodegrad. 125, 251–257 (2017).
Adams, R. I. et al. Ten questions concerning the microbiomes of buildings. Build. Environ. 109, 224–234 (2016).
Sundell, J. On the history of indoor air quality and health. Indoor Air 14 (Suppl. 7), 51–58 (2004).
Russell, F. A. R. The Atmosphere in Relation to Human Life and Health (Smithsonian Institution, 1896).
Carnelley, T., Haldane, J. S. & Anderson, A. M. The carbonic acid, organic matter, and micro-organisms in air, more especially of dwellings and schools. Phil. Trans. R. Soc. B Biol. Sci. 178, 61–111 (1887). This is a seminal study on indoor microorganisms — ahead of its time by about 100 years.
Carnelley, P. & Haldane, J. S. The air of sewers. Proc. R. Soc. 42, 394–396 (1887).
Carnelley, T. & Foggie, J. The air of schools. J. Pathol. Bacteriol. 2, 157–173 (1894).
Sedgwick, W. T. Principles of Sanitary Science and the. Public Health: With Special Reference to the Causation and Prevention of Infectious Diseases. (Macmillan, London, 1902).
Huddleson, I. F. & Hull, T. G. Bacteria of the air in an amusement hall. Am. J. Public Health N. Y. N. 1912 10, 583–585 (1920).
Sundell, J. et al. Ventilation rates and health: multidisciplinary review of the scientific literature: ventilation rates and health. Indoor Air 21, 191–204 (2011).
Quansah, R., Jaakkola, M. S., Hugg, T. T., Heikkinen, S. A. M. & Jaakkola, J. J. K. Residential dampness and molds and the risk of developing asthma: a systematic review and meta-analysis. PLOS ONE 7, e47526 (2012).
Carnelley, T. & Wilson, T. A. New method of determining the number of micro-organisms in air. Proc. R. Soc. 44, 455–464 (1888).
Graham-Smith, G. S. The microorganisms in the air of the House of Commons. J. Hyg. 3, 498–514 (1903).
Forbes, J. G. The atmosphere of the underground electric railways of London: a study of its bacterial content in 1920. J. Hyg. 22, 123–155 (1923).
Luckiesh, M., Taylor, A. H. & Holladay, L. L. Sampling devices for air-borne bacteria. J. Bacteriol. 52, 55–65 (1946).
Williams, R. E. O. & Hirch, A. The detection of streptococci in air. J. Hyg. 48, 504–524 (1950).
Williams, R. E. O., Lidwell, O. M. & Hirch, A. The bacterial flora of the air of occupied rooms. J. Hyg. 54, 512–523 (1956).
Reid, D. D., Lidwell, O. M. & Williams, R. E. O. Counts of air-borne bacteria as indices of air hygiene. J. Hyg. 54, 524–532 (1956).
Swaebly, M. A. & Christensen, C. M. Molds in house dust, furniture stuffing, and in the air within homes. J. Allergy 23, 370–374 (1952).
Buchbinder, L., Solowey, M. & Solotorovsky, M. Alpha hemolytic streptococci of air: their variant forms, origin and numbers per cubic foot of air in several types of locations. Am. J. Publ. Health Nat. Health 28, 61–71 (1938).
Christensen, C. M. Intramural dissemination of spores of Hormodendrum resinae. J. Allergy 21, 409–413 (1950).
Lidwell, O. M. & Lowbury, E. J. The survival of bacteria in dust. I. The distribution of bacteria in floor dust. J. Hyg. 48, 6–20 (1950).
Caplan, H. Observations on the role of hospital blankets as reservoirs of infection. J. Hyg. 60, 401–410 (1962).
Lidwell, O. M. & Lowbury, E. J. The survival of bacteria in dust. II. The effect of atmospheric humidity on the survival of bacteria in dust. J. Hyg. 48, 21–27 (1950).
Lidwell, O. M. & Lowbury, E. J. The survival of bacteria in dust. III. The effect of light on the survival of bacteria in dust. J. Hyg. 48, 28–37 (1950).
Wright, J., Cruickshank, R. & Gunn, W. Control of dust-borne streptococcal infection in measles wards. Br. Med. J. 1, 611–614 (1944).
Nash, T. Physical aspects of air disinfection. J. Hyg. 49, 382–399 (1951).
Hollaender, A. Ultra-violet irradiation as a means of disinfection of air. Am. J. Publ. Health Nat. Health 33, 980–984 (1943).
Lidwell, O. M. & Lowbury, E. J. The survival of bacteria in dust. IV. Atmospheric humidity and the bactericidal action of ultra-violet irradiation. J. Hyg. 48, 38–43 (1950).
Schaffer, N., Seidmon, E. E. & Bruskin, S. The clinical evaluation of air-borne and house dust fungi in New Jersey. J. Allergy 24, 348–354 (1953).
Maunsell, K. Air-borne fungal spores before and after raising dust; sampling by sedimentation. Int. Arch. Allergy Appl. Immunol. 3, 93–102 (1952).
Maunsell, K. Concentration of airborne spores in dwellings under normal conditions and under repair. Int. Arch. Allergy Appl. Immunol. 5, 373–376 (1954).
Winslow, C. E. & Robinson, E. A. An Investigation of the extent of the bacterial pollution of the atmosphere by mouth-spray. Am. J. Publ. Hyg. 20, 566–569 (1910).
Du Buy, H., Arnold, F. A. & Olson, B. J. Studies on the air transmission of micro-organisms derived from the respiratory tract: Lactobacillus acidophilus as a test organism. Publ. Health Rep. 62, 1391–1413 (1947).
Bourdillon, R. B. & Lidwell, O. M. Sneezing and the spread of infection. Lancet 238, 365–367 (1941).
Hart, D. Role of the respiratory tract in contamination of air: a comparative study. Arch. Surg. 38, 788 (1939).
Torrey, J. C. & Lake, M. Streptococci in air as an indicator of nasopharyngeal contamination. JAMA 117, 1425 (1941).
Wells, W. F. On air-borne infection. Study II. droplets and droplet nuclei. Am. J. Epidemiol. 20, 611–618 (1934). This is a seminal study on the method of disease transmission by small and large droplets expelled by humans.
Acheson, F. & Hewitt, D. Spread of influenza in a factory. Br. J. Soc. Med. 6, 68–75 (1952).
Moschandreas, D. J., Pagilla, K. R. & Storino, L. V. Time and space uniformity of indoor bacteria concentrations in Chicago area residences. Aerosol. Sci. Technol. 37, 899–906 (2003).
Tsai, F. C. & Macher, J. M. Concentrations of airborne culturable bacteria in 100 large US office buildings from the BASE study. Indoor Air 15, 71–81 (2005).
Reponen, T., Nevalainen, A. & Raunemaa, T. Bioaerosol and particle mass levels and ventilation in finnish homes. Environ. Int. 15, 203–208 (1989).
Kodama, A. M. & McGee, R. I. Airborne microbial contaminants in indoor environments. naturally ventilated and air-conditioned homes. Arch. Environ. Health Int. J. 41, 306–311 (1986).
Burger, H. Bioaerosols: prevalence and health effects in the indoor environment. J. Allergy Clin. Immunol. 86, 687–701 (1990).
Kaeberlein, T. Isolating ‘uncultivable’ microorganisms in pure culture in a simulated natural environment. Science 296, 1127–1129 (2002).
Zengler, K. et al. Cultivating the uncultured. Proc. Natl Acad. Sci. USA 99, 15681–15686 (2002).
Pace, N. R. A. Molecular view of microbial diversity and the biosphere. Science 276, 734–740 (1997).
Amann, R. I., Ludwig, W. & Schleifer, K. H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169 (1995).
Jackrel, S. L., Owens, S. M., Gilbert, J. A. & Pfister, C. A. Identifying the plant-associated microbiome across aquatic and terrestrial environments: the effects of amplification method on taxa discovery. Mol. Ecol. Resour. 17, 931–942 (2017).
Hatzenpichler, R. & Orphan, V. J. in Hydrocarbon and Lipid Microbiology Protocols (eds McGenity, T. J., Timmis, K. N. & Nogales, B.) 145–157 (Springer, Berlin, 2015)
Henry, C. S. et al. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 28, 977–982 (2010).
Cardona, C., Weisenhorn, P., Henry, C. & Gilbert, J. A. Network-based metabolic analysis and microbial community modeling. Curr. Opin. Microbiol. 31, 124–131 (2016).
Committee on Microbiomes of the Built Environment. Microbiomes of the Built Environment: A Research Agenda for Indoor Microbiology, Human Health, and Buildings (National Academies Press, 2017).
Acknowledgements
J.A.G. and B.S. acknowledge funding from the Alfred P. Sloan Foundation (060115). J.A.G. acknowledges S. Lax, C. Cardona and A. Sharma for their help in compiling references and for education about aspects of specific research elements.
Reviewer information
Nature Reviews Microbiology thanks R. Adams, M. Hernandez and M. Täubel for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
J.A.G. and B.S. researched data for the article, made substantial contributions to discussions of the content, wrote the article and reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
PROBIOM — Towards a health-promoting indoor microbiome: https://www.researchgate.net/project/PROBIOM-Towards-a-health-promoting-indoor-microbiome
Rights and permissions
About this article
Cite this article
Gilbert, J.A., Stephens, B. Microbiology of the built environment. Nat Rev Microbiol 16, 661–670 (2018). https://doi.org/10.1038/s41579-018-0065-5
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-018-0065-5
This article is cited by
-
Sociobiome - Individual and neighborhood socioeconomic status influence the gut microbiome in a multi-ethnic population in the US
npj Biofilms and Microbiomes (2024)
-
Targeted Metagenomics Identification of Microbiome in Preschools exposed to air Pollutants and Their Association with Respiratory Health symptom, Allergy and Eczema
Air Quality, Atmosphere & Health (2024)
-
Evaluation of rRNA depletion methods for capturing the RNA virome from environmental surfaces
BMC Research Notes (2023)
-
RNA-based amplicon sequencing is ineffective in measuring metabolic activity in environmental microbial communities
Microbiome (2023)
-
Aquaculture ecosystem microbiome at the water-fish interface: the case-study of rainbow trout fed with Tenebrio molitor novel diets
BMC Microbiology (2023)