Key Points
-
The fragile X mental retardation protein (FMRP) is a heterogeneous nuclear ribonucleoprotein (hnRNP) that shuttles between the nucleus and the cytoplasm of neuronal cells.
-
FMRP is present in the cell body as well as at synapses, where it regulates mRNA translation. The fragile X mental retardation 1 gene (FMR1) mRNA is also dendritically localized, and both the mRNA and the protein show activity-dependent localization.
-
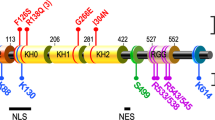
FMRP has four RNA-binding domains. It can recognize its target mRNAs directly or indirectly. Direct recognition occurs through an RNA structure called the G-quartet or through a poly(U) stretch. Indirect recognition occurs through the small non-coding RNA BC1 or microRNAs. Several hundreds of putative mRNA targets have been isolated so far.
-
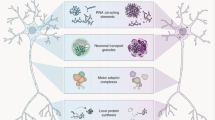
FMRP has different protein partners, the best characterized of which are those involved in mRNA transport and cytoskeleton structure.
-
Patients with fragile X syndrome and a mouse model of the syndrome both show learning and memory difficulties and anxiety. From a morphological point of view, neurons in patients and mice have longer, thinner dendritic spines. In vitro, the mouse model has been shown to have partial impairments in plasticity, such as long-term potentiation and long-term depression.
-
Recently, a new syndrome has been associated with the FMR1 gene — fragile X tremor ataxia syndrome. The clinical presentation includes either gait ataxia or intention tremor at onset, and is associated with a wide range of neurological symptoms and signs, including cognitive impairment, parkinsonism, peripheral neuropathy and autonomic dysfunction.
Abstract
The mental retardation protein FMRP is involved in the transport of mRNAs and their translation at synapses. Patients with fragile X syndrome, in whom FMRP is absent or mutated, show deficits in learning and memory that might reflect impairments in the translational regulation of a subset of neuronal mRNAs. The study of FMRP provides important insights into the regulation and functions of local protein synthesis in the neuronal periphery, and increases our understanding of how these functions can produce specific effects at individual synapses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Oberle, I. et al. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science 252, 1097–1102 (1991).
Hagerman, P. J. & Hagerman, R. J. Fragile X-associated tremor/ataxia syndrome (FXTAS). Ment. Retard. Dev. Disabil. Res. Rev. 10, 25–30 (2004).
Hinton, V. J., Brown, W. T., Wisniewski, K. & Rudelli, R. D. Analysis of neocortex in three males with the fragile X syndrome. Am. J. Med. Genet. 41, 289–294 (1991).
Comery, T. A. et al. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc. Natl Acad. Sci. USA 94, 5401–5404 (1997).
Irwin, S. A. et al. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am. J. Med. Genet. 98, 161–167 (2001). Provides the most thorough and the only quantitative description of the human fragile X cortical neuronal morphological phenotype.
Nimchinsky, E. A., Oberlander, A. M. & Svoboda, K. Abnormal development of dendritic spines in FMR1 knock-out mice. J. Neurosci. 21, 5139–5146 (2001).
Greenough, W. T. et al. Synaptic regulation of protein synthesis and the fragile X protein. Proc. Natl Acad. Sci. USA 98, 7101–7106 (2001).
Coss, R. G. & Perkel, D. H. The function of dendritic spines: a review of theoretical issues. Behav. Neural Biol. 44, 151–185 (1985).
Nimchinsky, E. A., Sabatini, B. L. & Svoboda, K. Structure and function of dendritic spines. Annu. Rev. Physiol. 64, 313–353 (2002).
Campbell, D. S. & Holt, C. E. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron 32, 1013–1026 (2001).
Steward, O. & Schuman, E. M. Compartmentalized synthesis and degradation of proteins in neurons. Neuron 40, 347–359 (2003).
Van de Bor, V. & Davis, I. mRNA localisation gets more complex. Curr. Opin. Cell Biol. 16, 300–307 (2004).
Farina, K. L. & Singer, R. H. The nuclear connection in RNA transport and localization. Trends Cell Biol. 12, 466–472 (2002).
Gu, W., Pan, F., Zhang, H., Bassell, G. J. & Singer, R. H. A predominantly nuclear protein affecting cytoplasmic localization of β-actin mRNA in fibroblasts and neurons. J. Cell Biol. 156, 41–51 (2002).
Ainger, K. et al. Transport and localization elements in myelin basic protein mRNA. J. Cell Biol. 138, 1077–1087 (1997).
Shan, J., Munro, T. P., Barbarese, E., Carson, J. H. & Smith, R. A molecular mechanism for mRNA trafficking in neuronal dendrites. J. Neurosci. 23, 8859–8866 (2003).
Deshler, J. O., Highett, M. I. & Schnapp, B. J. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science 276, 1128–1131 (1997).
Ross, A. F., Oleynikov, Y., Kislauskis, E. H., Taneja, K. L. & Singer, R. H. Characterization of a β-actin mRNA zipcode-binding protein. Mol. Cell Biol. 17, 2158–2165 (1997).
Tiruchinapalli, D. M. et al. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and β-actin mRNA in dendrites and spines of hippocampal neurons. J. Neurosci. 23, 3251–3261 (2003).
Kislauskis, E. H., Zhu, X. & Singer, R. H. Sequences responsible for intracellular localization of β-actin messenger RNA also affect cell phenotype. J. Cell Biol. 127, 441–451 (1994).
Palacios, I. M., Gatfield, D., St Johnston, D. & Izaurralde, E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427, 753–757 (2004). Shows that the translation-initiation factor eIF4AIII is a component of the oskar mRNP localization complex in D. melanogaster . Moreover, the same factor is also involved in nonsense-mediated mRNA decay in mammals.
Singh, G. & Lykke-Andersen, J. New insights into the formation of active nonsense-mediated decay complexes. Trends Biochem. Sci. 28, 464–466 (2003).
Holbrook, J. A., Neu-Yilik, G., Hentze, M. W. & Kulozik, A. E. Nonsense-mediated decay approaches the clinic. Nature Genet. 36, 801–808 (2004).
Weiler, I. J. et al. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc. Natl Acad. Sci. USA 94, 5395–5400 (1997). Initial description of neurotransmitter activation of FMRP synthesis at the synapse (see also reference 25).
Weiler, I. J. et al. From the cover: fragile X mental retardation protein is necessary for neurotransmitter-activated protein translation at synapses. Proc. Natl Acad. Sci. USA 101, 17504–17509 (2004).
Verheij, C. et al. Characterization and localization of the FMR-1 gene product associated with fragile X syndrome. Nature 363, 722–724 (1993).
Eberhart, D. E., Malter, H. E., Feng, Y. & Warren, S. T. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum. Mol. Genet. 5, 1083–1091 (1996). An initial, detailed characterization of FMRP.
Feng, Y. et al. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J. Neurosci. 17, 1539–1547 (1997).
Khandjian, E. W. et al. Novel isoforms of the fragile X related protein FXR1P are expressed during myogenesis. Hum. Mol. Genet. 7, 2121–2128 (1998).
Ceman, S., Brown, V. & Warren, S. T. Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex. Mol. Cell Biol. 19, 7925–7932 (1999).
Bardoni, B. et al. NUFIP1 (nuclear FMRP interacting protein 1) is a nucleocytoplasmic shuttling protein associated with active synaptoneurosomes. Exp. Cell Res. 289, 95–107 (2003).
Ceman, S., Nelson, R. & Warren, S. T. Identification of mouse YB1/p50 as a component of the FMRP-associated mRNP particle. Biochem. Biophys. Res. Commun. 279, 904–908 (2000).
Stickeler, E. et al. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 20, 3821–3830 (2001).
Nekrasov, M. P. et al. The mRNA-binding protein YB-1 (p50) prevents association of the eukaryotic initiation factor eIF4G with mRNA and inhibits protein synthesis at the initiation stage. J. Biol. Chem. 278, 13936–13943 (2003).
Sittler, A., Devys, D., Weber, C. & Mandel, J. L. Alternative splicing of exon 14 determines nuclear or cytoplasmic localisation of fmr1 protein isoforms. Hum. Mol. Genet. 5, 95–102 (1996).
De Boulle, K. et al. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nature Genet. 3, 31–35 (1993).
Tamanini, F. et al. Different targets for the fragile X-related proteins revealed by their distinct nuclear localizations. Hum. Mol. Genet. 8, 863–869 (1999).
Nakamura, M. et al. When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to γ-tubulin. J. Cell Biol. 143, 1041–1052 (1998).
Menon, R. P., Gibson, T. J. & Pastore, A. The C terminus of fragile X mental retardation protein interacts with the multi-domain Ran-binding protein in the microtubule-organising centre. J. Mol. Biol. 343, 43–53 (2004).
Hoelz, A. & Blobel, G. Cell biology: popping out of the nucleus. Nature 432, 815–816 (2004).
Greenbaum, L., Katcoff, D. J., Dou, H., Gozlan, Y. & Malik, Z. A porphobilinogen deaminase (PBGD) Ran-binding protein interaction is implicated in nuclear trafficking of PBGD in differentiating glioma cells. Oncogene 22, 5221–5228 (2003).
Ashley, C. T. et al. Human and murine FMR-1: alternative splicing and translational initiation downstream of the CGG-repeat. Nature Genet. 4, 244–251 (1993).
Zalfa, F. et al. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 112, 317–327 (2003). This work reveals an intriguing mechanism by which FMRP regulates the translation of some neuronal mRNAs at synapses through the small non-coding RNA BC1. This RNA functions as an 'adaptor molecule' between FMRP and its mRNA targets. The FMRP–mRNA target interaction can be inhibited using a chemically synthesized oligo against BC1 RNA.
Jin, P. et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nature Neurosci. 7, 113–117 (2004).
He, L. & Hannon, G. J. MicroRNAs: small RNAs with a big role in gene regulation. Nature Rev. Genet. 5, 522–531 (2004).
Jenuwein, T. Molecular biology. An RNA-guided pathway for the epigenome. Science 297, 2215–2218 (2002).
Bao, N., Lye, K. W. & Barton, M. K. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell 7, 653–662 (2004).
Matzke, M. A. & Birchler, J. A. RNAi-mediated pathways in the nucleus. Nature Rev. Genet. 6, 24–35 (2005).
Antar, L. N., Afroz, R., Dictenberg, J. B., Carroll, R. C. & Bassell, G. J. Metabotropic glutamate receptor activation regulates fragile X mental retardation protein and Fmr1 mRNA localization differentially in dendrites and at synapses. J. Neurosci. 24, 2648–2655 (2004). Shows clearly, for the first time, evidence of activity-dependent regulation of FMRP and FMR1 mRNA trafficking in dendrites and synapses. In particular, the authors show that synaptic activation through the mGluR5 receptor regulates the localization of FMRP and FMR1 mRNA in dendrites.
Miyashiro, K. Y. et al. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron 37, 417–431 (2003).
Steward, O., Bakker, C. E., Willems, P. J. & Oostra, B. A. No evidence for disruption of normal patterns of mRNA localization in dendrites or dendritic transport of recently synthesized mRNA in FMR1 knockout mice, a model for human fragile-X mental retardation syndrome. Neuroreport 9, 477–481 (1998).
Kanai, Y., Dohmae, N. & Hirokawa, N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525 (2004). A thorough description and functional assessment of the composition of granules that contain FMRP and transport its mRNAs (see also references 49,60). They identified more than 40 proteins including Staufen, FMRP and other factors involved in protein synthesis. In addition, they showed the presence of α-CaMKII and ARC mRNAs in these granules.
Rackham, O. & Brown, C. M. Visualization of RNA–protein interactions in living cells: FMRP and IMP1 interact on mRNAs. EMBO J. 23, 3346–3355 (2004).
Ling, S. C., Fahrner, P. S., Greenough, W. T. & Gelfand, V. I. Transport of Drosophila fragile X mental retardation protein-containing ribonucleoprotein granules by kinesin-1 and cytoplasmic dynein. Proc. Natl Acad. Sci. USA 101, 17428–17433 (2004).
Schrier, M. et al. Transport kinetics of FMRP containing the I304N mutation of severe fragile X syndrome in neurites of living rat PC12 cells. Exp. Neurol. 189, 343–353 (2004).
Clarke, N. F., Mowat, D., Kooy, R. F., Reyniers, E. & Willemsen, R. Fragile X syndrome phenotype with normal FMR1 gene studies. Am. J. Med. Genet. A. 129, 326–328 (2004).
Zhang, Y. et al. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 14, 5358–5366 (1995).
Schenck, A., Bardoni, B., Moro, A., Bagni, C. & Mandel, J. L. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc. Natl Acad. Sci. USA 98, 8844–8849 (2001). The authors characterized two cytoplasmic FMRP-interacting proteins, both of which are also present at synapses.This work represents one of the first links between FMRP and the cytoskeleton (see also reference 68).
Bardoni, B. & Mandel, J. L. Advances in understanding of fragile X pathogenesis and FMRP function, and in identification of X linked mental retardation genes. Curr. Opin. Genet. Dev. 12, 284–293 (2002).
Ohashi, S. et al. Identification of mRNA/protein (mRNP) complexes containing Purα, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J. Biol. Chem. 277, 37804–37810 (2002). Shows that a possible molecular machinery containing PURα, Staufen, myosin VA and FMRP might be involved in the regulatation of dendritic transport.
Villace, P., Marion, R. M. & Ortin, J. The composition of Staufen-containing RNA granules from human cells indicates their role in the regulated transport and translation of messenger RNAs. Nucleic Acids Res. 32, 2411–2420 (2004).
Siomi, M. C., Zhang, Y., Siomi, H. & Dreyfuss, G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol. Cell Biol. 16, 3825–3832 (1996).
Adinolfi, S. et al. The N-terminus of the fragile X mental retardation protein contains a novel domain involved in dimerization and RNA binding. Biochemistry 42, 10437–10444 (2003).
Agulhon, C. et al. Expression of FMR1, FXR1, and FXR2 genes in human prenatal tissues. J. Neuropathol. Exp. Neurol. 58, 867–880 (1999).
Sreeram, N., Wren, C., Bhate, M., Robertson, P. & Hunter, S. Cardiac abnormalities in the fragile X syndrome. Br. Heart J. 61, 289–291 (1989).
Mientjes, E. J. et al. Fxr1 knockout mice show a striated muscle phenotype: implications for Fxr1p function in vivo. Hum. Mol. Genet. 13, 1291–1302 (2004).
Kobayashi, K. et al. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J. Biol. Chem. 273, 291–295 (1998).
Schenck, A. et al. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron 38, 887–898 (2003).
Bardoni, B. et al. 82-FIP, a novel FMRP (fragile X mental retardation protein) interacting protein, shows a cell cycle-dependent intracellular localization. Hum. Mol. Genet. 12, 1689–1698 (2003).
Kohrmann, M. et al. Microtubule-dependent recruitment of Staufen–green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Biol. Cell 10, 2945–2953 (1999).
Li, Y. et al. Pur α protein implicated in dendritic RNA transport interacts with ribosomes in neuronal cytoplasm. Biol. Pharm. Bull. 24, 231–235 (2001).
Duchaine, T. F. et al. Staufen2 isoforms localize to the somatodendritic domain of neurons and interact with different organelles. J. Cell Sci. 115, 3285–3295 (2002).
Ohashi, S. et al. The single-stranded DNA- and RNA-binding proteins pur α and pur β link BC1 RNA to microtubules through binding to the dendrite-targeting RNA motifs. J. Neurochem. 75, 1781–1790 (2000).
Mallardo, M. et al. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc. Natl Acad. Sci. USA 100, 2100–2105 (2003).
Pastural, E. et al. Two genes are responsible for Griscelli syndrome at the same 15q21 locus. Genomics 63, 299–306 (2000).
Siomi, M. C. et al. FXR1, an autosomal homolog of the fragile X mental retardation gene. EMBO J. 14, 2401–2408 (1995).
Adinolfi, S. et al. Dissecting FMR1, the protein responsible for fragile X syndrome, in its structural and functional domains. RNA 5, 1248–1258 (1999).
Brown, V. et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107, 477–487 (2001).
Darnell, J. C. et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 107, 489–499 (2001). Describes an approach to identifying mRNA 'cargoes' of FMRP (see also references 50,78,80).
Chen, L., Yun, S. W., Seto, J., Liu, W. & Toth, M. The fragile X mental retardation protein binds and regulates a novel class of mRNAs containing U rich target sequences. Neuroscience 120, 1005–1017 (2003).
Zhang, Y. Q. et al. Drosophila fragile X-related gene regulates the MAP1B homolog futsch to control synaptic structure and function. Cell 107, 591–603 (2001).
Schaeffer, C. et al. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 20, 4803–4813 (2001).
Hessl, D., Rivera, S. M. & Reiss, A. L. The neuroanatomy and neuroendocrinology of fragile X syndrome. Ment. Retard. Dev. Disabil. Res. Rev. 10, 17–24 (2004).
Ramos, A., Hollingworth, D. & Pastore, A. G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA 9, 1198–1207 (2003).
Rozhdestvensky, T. S., Kopylov, A. M., Brosius, J. & Huttenhofer, A. Neuronal BC1 RNA structure: evolutionary conversion of a tRNA(Ala) domain into an extended stem-loop structure. RNA 7, 722–730 (2001).
Gabus, C., Mazroui, R., Tremblay, S., Khandjian, E. W. & Darlix, J. L. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Res. 32, 2129–2137 (2004).
Caudy, A. A., Myers, M., Hannon, G. J. & Hammond, S. M. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 16, 2491–2496 (2002). This study provides the first evidence that the D. melanogaster FMRP is part of the RNAi-related machinery (see also reference 88).
Ishizuka, A., Siomi, M. C. & Siomi, H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 16, 2497–2508 (2002). The authors of this study suggest a model in which the RNAi pathway and D. melanogaster FMRP-mediated translational control intersect with each other (see also reference 86).
Gebauer, F. & Hentze, M. W. Molecular mechanisms of translational control. Nature Rev. Mol. Cell Biol. 5, 827–835 (2004).
Chapman, R. E. & Walter, P. Translational attenuation mediated by an mRNA intron. Curr. Biol. 7, 850–859 (1997).
Kedersha, N. & Anderson, P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30, 963–969 (2002).
Krichevsky, A. M. & Kosik, K. S. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron 32, 683–696 (2001).
Antic, D. & Keene, J. D. Messenger ribonucleoprotein complexes containing human ELAV proteins: interactions with cytoskeleton and translational apparatus. J. Cell Sci. 111, 183–197 (1998).
Khandjian, E. W., Corbin, F., Woerly, S. & Rousseau, F. The fragile X mental retardation protein is associated with ribosomes. Nature Genet. 12, 91–93 (1996).
Stefani, G., Fraser, C. E., Darnell, J. C. & Darnell R. B. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J. Neurosci. 24, 7272–7276 (2004).
Ceman, S. et al. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum. Mol. Genet. 12, 3295–3305 (2003).
Aschrafi, A., Cunningham, B. A., Edelman, G. M. & Vanderklish, P. W. The fragile X mental retardation protein and group I metabotropic glutamate receptors regulate levels of mRNA granules in brain. Proc. Natl Acad. Sci. USA 102, 2180–2185 (2005).
Purpura, D. P. Dendritic spine 'dysgenesis' and mental retardation. Science 186, 1126–1128 (1974).
Marin-Padilla, M. Structural abnormalities of the cerebral cortex in human chromosomal aberrations: a Golgi study. Brain Res. 44, 625–629 (1972).
Wisniewski, K. E. et al. Fragile X syndrome: associated neurological abnormalities and developmental disabilities. Ann. Neurol. 18, 665–669 (1985).
Rudelli, R. D. et al. Adult fragile X syndrome. Clinico-neuropathologic findings. Acta Neuropathol. (Berl.) 67, 289–295 (1985).
McKinney, B. C., Grossman, A. W., Elisseou, N. M. & Greenough, W. T. Dendritic spine abnormalities in the occipital cortex of C57BL/6 Fmr1 knockout mice. Am. J. Med. Genet. (in the press).
Bakker, C. E. et al. Fmr1 knockout mice: a model to study fragile X mental retardation. Cell 78, 23–33 (1994).
Braun, K. & Segal, M. FMRP involvement in formation of synapses among cultured hippocampal neurons. Cereb. Cortex 10, 1045–1052 (2000).
Galvez, R. & Greenough, W. T. Sequence of abnormal dendritic spine development in primary somatosensory cortex of a mouse model of the fragile X mental retardation syndrome. Am. J. Med. Genet. (in the press).
Greenough, W. T. & Chang, F. L. Dendritic pattern formation involves both oriented regression and oriented growth in the barrels of mouse somatosensory cortex. Brain Res. 471, 148–152 (1988).
Huttenlocher, P. R. & Dabholkar, A. S. Regional differences in synaptogenesis in human cerebral cortex. J. Comp. Neurol. 387, 167–178 (1997).
Galvez, R., Gopal, A. R. & Greenough, W. T. Somatosensory cortical barrel dendritic abnormalities in a mouse model of the fragile X mental retardation syndrome. Brain Res. 971, 83–89 (2003). Evidence that synaptic pruning deficiencies give rise to the neuronal phenotype of fragile X syndrome.
Volkmar, F. R. & Greenough, W. T. Rearing complexity affects branching of dendrites in the visual cortex of the rat. Science 176, 1145–1147 (1972).
van Praag, H., Kempermann, G. & Gage, F. H. Neural consequences of environmental enrichment. Nature Rev. Neurosci. 1, 191–198 (2000).
Benaroya-Milshtein, N. et al. Environmental enrichment in mice decreases anxiety, attenuates stress responses and enhances natural killer cell activity. Eur. J. Neurosci. 20, 1341–1347 (2004).
Schrijver, N. C., Bahr, N. I., Weiss, I. C. & Wurbel, H. Dissociable effects of isolation rearing and environmental enrichment on exploration, spatial learning and HPA activity in adult rats. Pharmacol. Biochem. Behav. 73, 209–224 (2002).
Moser, M. B., Trommald, M., Egeland, T. & Andersen, P. Spatial training in a complex environment and isolation alter the spine distribution differently in rat CA1 pyramidal cells. J. Comp. Neurol. 380, 373–381 (1997).
Lee, E. H., Hsu, W. L., Ma, Y. L., Lee, P. J. & Chao, C. C. Enrichment enhances the expression of sgk, a glucocorticoid-induced gene, and facilitates spatial learning through glutamate AMPA receptor mediation. Eur. J. Neurosci. 18, 2842–2852 (2003).
Greenough, W. T., Yuwiler, A. & Dollinger, M. Effects of posttrial eserine administration on learning in 'enriched'- and 'impoverished'-reared rats. Behav. Biol. 8, 261–272 (1973).
Diamond, M. C. & Connor, J. R. Jr. Plasticity of the aging cerebral cortex. Exp. Brain Res. 5 (suppl.), 36–44 (1982).
Turner, A. M. & Greenough, W. T. Differential rearing effects on rat visual cortex synapses. I. Synaptic and neuronal density and synapses per neuron. Brain Res. 329, 195–203 (1985).
Irwin, S. A. et al. Evidence for altered Fragile-X mental retardation protein expression in response to behavioral stimulation. Neurobiol. Learn. Mem. 74, 87–93 (2000).
Todd, P. K. & Mack, K. J. Sensory stimulation increases cortical expression of the fragile X mental retardation protein in vivo. Brain Res. Mol. Brain Res. 80, 17–25 (2000).
Ostroff, L. E., Fiala, J. C., Allwardt, B. & Harris, K. M. Polyribosomes redistribute from dendritic shafts into spines with enlarged synapses during LTP in developing rat hippocampal slices. Neuron 35, 535–545 (2002). A landmark study on how dendritic protein synthesis might stabilize the growth of the postsynaptic density, including an increase in the number of ribosomes, after the induction of LTP by tetanic stimulation.
Chang, F. L. & Greenough, W. T. Transient and enduring morphological correlates of synaptic activity and efficacy change in the rat hippocampal slice. Brain Res. 309, 35–46 (1984).
Greenough, W. T., Hwang, H. M. & Gorman, C. Evidence for active synapse formation or altered postsynaptic metabolism in visual cortex of rats reared in complex environments. Proc. Natl Acad. Sci. USA 82, 4549–4552 (1985).
Huber, K. M., Gallagher, S. M., Warren, S. T. & Bear, M. F. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc. Natl Acad. Sci. USA 99, 7746–7750 (2002).
Li, J., Pelletier, M. R., Perez Velazquez, J. L. & Carlen, P. L. Reduced cortical synaptic plasticity and GluR1 expression associated with fragile X mental retardation protein deficiency. Mol. Cell Neurosci. 19, 138–151 (2002).
Snyder, E. M. et al. Internalization of ionotropic glutamate receptors in response to mGluR activation. Nature Neurosci. 4, 1079–1085 (2001).
Vanderklish, P. W. & Edelman, G. M. Dendritic spines elongate after stimulation of group 1 metabotropic glutamate receptors in cultured hippocampal neurons. Proc. Natl Acad. Sci. USA 99, 1639–1644 (2002).
Bear, M. F., Huber, K. M. & Warren, S. T. The mGluR theory of fragile X mental retardation. Trends Neurosci. 27, 370–377 (2004).
Godfraind, J. M. et al. Long-term potentiation in the hippocampus of fragile X knockout mice. Am. J. Med. Genet. 64, 246–251 (1996). Initial evidence for a direct effect of FMR1 knockout on a widely studied model of learning and memory (see also reference 122).
Siomi, H., Siomi, M. C., Nussbaum, R. L. & Dreyfuss, G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell 74, 291–298 (1993).
Adinolfi, S. et al. Novel RNA-binding motif: the KH module. Biopolymers 51, 153–164 (1999).
Gibson, T. J., Thompson, J. D. & Heringa, J. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 324, 361–366 (1993).
Lewis, H. A. et al. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell 100, 323–332 (2000).
Duncan, R. et al. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 8, 465–480 (1994).
Michelotti, E. F., Michelotti, G. A., Aronsohn, A. I. & Levens, D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell Biol. 16, 2350–2360 (1996).
Kiledjian, M. & Dreyfuss, G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 11, 2655–2664 (1992).
Zhang, S. & Grosse, F. Domain structure of human nuclear DNA helicase II (RNA helicase A). J. Biol. Chem. 272, 11487–11494 (1997).
Sandri-Goldin, R. M. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12, 868–879 (1998).
Ghisolfi, L., Kharrat, A., Joseph, G., Amalric, F. & Erard, M. Concerted activities of the RNA recognition and the glycine-rich C-terminal domains of nucleolin are required for efficient complex formation with pre-ribosomal RNA. Eur. J. Biochem. 209, 541–548 (1992).
Fouraux, M. A., Bouvet, P., Verkaart, S., van Venrooij, W. J. & Pruijn, G. J. Nucleolin associates with a subset of the human Ro ribonucleoprotein complexes. J. Mol. Biol. 320, 475–488 (2002).
Lapeyre, B. et al. Molecular cloning of Xenopus fibrillarin, a conserved U3 small nuclear ribonucleoprotein recognized by antisera from humans with autoimmune disease. Mol. Cell Biol. 10, 430–434 (1990).
Bagni, C. & Lapeyre, B. Gar1p binds to the small nucleolar RNAs snR10 and snR30 in vitro through a nontypical RNA binding element. J. Biol. Chem. 273, 10868–10873 (1998).
Lee, W. C., Xue, Z. X. & Melese, T. The NSR1 gene encodes a protein that specifically binds nuclear localization sequences and has two RNA recognition motifs. J. Cell Biol. 113, 1–12 (1991).
Nichols, R. C. et al. The RGG domain in hnRNP A2 affects subcellular localization. Exp. Cell Res. 256, 522–532 (2000).
Maurer-Stroh, S. et al. The Tudor domain 'Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 28, 69–74 (2003).
Siomi, H., Choi, M., Siomi, M. C., Nussbaum, R. L. & Dreyfuss, G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell 77, 33–39 (1994).
Feng, Y. et al. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell 1, 109–118 (1997).
Brown, V. et al. Purified recombinant Fmrp exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein. J. Biol. Chem. 273, 15521–15527 (1998).
Laggerbauer, B., Ostareck, D., Keidel, E. M., Ostareck-Lederer, A. & Fischer, U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum. Mol. Genet. 10, 329–338 (2001).
Li, Z. et al. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 29, 2276–2283 (2001).
Hollmann, M. & Heinemann, S. Cloned glutamate receptors. Annu. Rev. Neurosci. 17, 31–108 (1994).
Bardoni, B., Schenck, A. & Mandel, J. L. A novel RNA-binding nuclear protein that interacts with the fragile X mental retardation (FMR1) protein. Hum. Mol. Genet. 8, 2557–2566 (1999).
Brendel, C. et al. Characterization of Staufen 1 ribonucleoprotein complexes. Biochem. J. 384, 239–246 (2004).
Acknowledgements
We are grateful to T. Achsel, I. J. Weiler and F. Zalfa for critical reading of the manuscript and to G. Bernardi for his support. Work in C.B.'s group was supported by Telethon-Italy, Ministero dell'Istruzione, dell'Università e della Ricerca and Ministero della Salute. Work in W.G.'s laboratory was supported by the National Institute of Mental Health, the National Institute of Child Health and Human Developent and the Fragile X Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez Gene
OMIM
FURTHER INFORMATION
The Fragile X Research Foundation
Glossary
- CpG ISLANDS
-
Sequences of at least 200 bp with greater than 50% G+C content and high CpG frequency.
- G-QUARTETS
-
Intramolecular, bimolecular or tetramolecular structures. Guanine is arranged in series to form a planar layer, which is cation dependent.
- NONSENSE-MEDIATED DECAY
-
(NMD). A mechanism by which cells recognize and degrade mRNAs that prematurely terminate translation.
- MESSENGER RIBONUCLEOPROTEIN
-
(mRNP). An mRNA, associated with proteins, that is translationally inactive.
- K HOMOLOGY DOMAINS
-
(KH domain). A sequence motif that was originally identified as three repeats in the human hnRNP K protein and that is present in proteins from bacteria through to humans. The motif expands around a conserved VIGXXGXXI core (where X is any amino acid, with a preference for positive residues).
- RGG BOX
-
A protein region that is rich in arginine and glycine residues, is positively charged and has a high affinity for RNA molecules.
- CHROMATIN REMODELLING
-
Epigenetic DNA modifications that silence genes at a transcriptional level without altering their structure. Alterations in chromatin remodelling cause various multi-system disorders and neoplasias.
- microRNA
-
A non-coding RNA molecule of 21–24 nucleotides that inhibits mRNA expression.
- SYNAPTONEUROSOMES
-
Purified synapses containing the pre- and postsynaptic termini. The presynaptic compartment contains the synaptic vesicles and the postsynaptic compartment contains the translational machinery.
- LYMPHOBLAST CELLS
-
Immortalized blood cells (lymphocytes).
- POLYSOME
-
A string of 80S ribosomes bound to an mRNA molecule.
- ENVIRONMENTAL ENRICHMENT
-
A combination of complex inanimate and social stimulation. Toys, ladders, tunnels and a running wheel are placed in the cage for voluntary physical exercise and are routinely changed during experimental periods.
Rights and permissions
About this article
Cite this article
Bagni, C., Greenough, W. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci 6, 376–387 (2005). https://doi.org/10.1038/nrn1667
Issue Date:
DOI: https://doi.org/10.1038/nrn1667
This article is cited by
-
EF1α-associated protein complexes affect dendritic spine plasticity by regulating microglial phagocytosis in Fmr1 knock-out mice
Molecular Psychiatry (2024)
-
Tau reduction attenuates autism-like features in Fmr1 knockout mice
Molecular Autism (2023)
-
Caveolin-1-Mediated Cholesterol Accumulation Contributes to Exaggerated mGluR-Dependent Long-Term Depression and Impaired Cognition in Fmr1 Knockout Mice
Molecular Neurobiology (2023)
-
Dendrites of Neocortical Pyramidal Neurons: The Key to Understand Intellectual Disability
Cellular and Molecular Neurobiology (2022)
-
Increased 2-arachidonoyl-sn-glycerol levels normalize cortical responses to sound and improve behaviors in Fmr1 KO mice
Journal of Neurodevelopmental Disorders (2021)