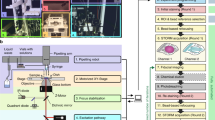
Abstract
Single-molecule localization microscopy (SMLM) is rapidly gaining popularity in the life sciences as an efficient approach to visualize molecular distribution with nanoscale precision. However, it has been challenging to obtain and analyze such data within a cellular context in tissue preparations. Here we describe a 5-d tissue processing and immunostaining procedure that is optimized for SMLM, and we provide example applications to fixed mouse brain, heart and kidney tissues. We then describe how to perform correlated confocal and 3D-superresolution imaging on these sections, which allows the visualization of nanoscale protein localization within labeled subcellular compartments of identified target cells in a few minutes. Finally, we describe the use of VividSTORM (http://katonalab.hu/index.php/vividstorm), an open-source software for correlated confocal and SMLM image analysis, which facilitates the measurement of molecular abundance, clustering, internalization, surface density and intermolecular distances in a cell-specific and subcellular compartment–restricted manner. The protocol requires only basic skills in tissue staining and microscopy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
References
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Hess, S.T., Girirajan, T.P. & Mason, M.D. Ultra-high-resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 4258–4272 (2006).
Rust, M.J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006).
Sauer, M. Localization microscopy coming of age: from concepts to biological impact. J. Cell Sci. 126, 3505–3513 (2013).
Dudok, B. et al. Cell-specific STORM super-resolution imaging reveals nanoscale organization of cannabinoid signaling. Nat. Neurosci. 18, 75–86 (2015).
Usoskin, D. et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 18, 145–153 (2015).
Zeisel, A. et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015).
Darmanis, S. et al. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. USA 112, 7285–7290 (2015).
Klausberger, T. & Somogyi, P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science 321, 53–57 (2008).
O'Rourke, N.A., Weiler, N.C., Micheva, K.D. & Smith, S.J. Deep molecular diversity of mammalian synapses: why it matters and how to measure it. Nat. Rev. Neurosci. 13, 365–379 (2012).
Choquet, D. & Triller, A. The dynamic synapse. Neuron 80, 691–703 (2013).
Ludányi, A. et al. Downregulation of the CB1 cannabinoid receptor and related molecular elements of the endocannabinoid system in epileptic human hippocampus. J. Neurosci. 28, 2976–2990 (2008).
Jung, K.M. et al. Uncoupling of the endocannabinoid signalling complex in a mouse model of fragile X syndrome. Nat. Commun. 3, 1080 (2012).
Nusser, Z. A new approach to estimate the number, density and variability of receptors at central synapses. Eur. J. Neurosci. 11, 745–752 (1999).
Masugi-Tokita, M. & Shigemoto, R. High-resolution quantitative visualization of glutamate and GABA receptors at central synapses. Curr. Opin. Neurobiol. 17, 387–393 (2007).
Requejo-Isidro, J. Fluorescence nanoscopy. Methods and applications. J. Chem. Biol. 6, 97–120 (2013).
Yamanaka, M., Smith, N.I. & Fujita, K. Introduction to super-resolution microscopy. Microscopy (Oxf) 63, 177–192 (2014).
Maglione, M. & Sigrist, S.J. Seeing the forest tree by tree: super-resolution light microscopy meets the neurosciences. Nat. Neurosci. 16, 790–797 (2013).
Oddone, A., Vilanova, I.V., Tam, J. & Lakadamyali, M. Super-resolution imaging with stochastic single-molecule localization: concepts, technical developments, and biological applications. Microsc. Res. Tech. 77, 502–509 (2014).
Lee, S.H. et al. Multiple forms of endocannabinoid and endovanilloid signaling regulate the tonic control of GABA release. J. Neurosci. 35, 10039–10057 (2015).
Dani, A., Huang, B., Bergan, J., Dulac, C. & Zhuang, X. Superresolution imaging of chemical synapses in the brain. Neuron 68, 843–856 (2010).
Heilemann, M. et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 47, 6172–6176 (2008).
Godin, A.G., Lounis, B. & Cognet, L. Super-resolution microscopy approaches for live cell imaging. Biophys. J. 107, 1777–1784 (2014).
Dempsey, G.T., Vaughan, J.C., Chen, K.H., Bates, M. & Zhuang, X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods 8, 1027–1036 (2011).
Bates, M., Huang, B., Dempsey, G.T. & Zhuang, X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 317, 1749–1753 (2007).
Lakadamyali, M. Super-resolution microscopy: going live and going fast. Chemphyschem 15, 630–636 (2014).
Jones, S.A., Shim, S.H., He, J. & Zhuang, X. Fast, three-dimensional super-resolution imaging of live cells. Nat. Methods 8, 499–508 (2011).
Shim, S.H. et al. Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc. Natl. Acad. Sci. USA 109, 13978–13983 (2012).
Zhong, G. et al. Developmental mechanism of the periodic membrane skeleton in axons. Elife 3 http://dx.doi.org/10.7554/eLife.04581 (23 December 2014).
van de Linde, S. et al. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 6, 991–1009 (2011).
Gould, T.J., Verkhusha, V.V. & Hess, S.T. Imaging biological structures with fluorescence photoactivation localization microscopy. Nat. Protoc. 4, 291–308 (2009).
MacGillavry, H.D. & Blanpied, T.A. Single-molecule tracking photoactivated localization microscopy to map nano-scale structure and dynamics in living spines. Curr. Protoc. Neurosci. 2, 2.20.1–2.20.19 (2013).
Löschberger, A., Franke, C., Krohne, G., van de Linde, S. & Sauer, M. Correlative super-resolution fluorescence and electron microscopy of the nuclear pore complex with molecular resolution. J. Cell Sci. 127, 4351–4355 (2014).
Tam, J. et al. A microfluidic platform for correlative live-cell and super-resolution microscopy. PLoS ONE 9, e115512 (2014).
Tam, J. & Merino, D. STORM in comparison with STED and other imaging methods. J. Neurochem. 135, 643–658 (2015).
Tønnesen, J. & Nägerl, U.V. Superresolution imaging for neuroscience. Exp. Neurol. 242, 33–40 (2013).
Specht, C.G. et al. Quantitative nanoscopy of inhibitory synapses: counting gephyrin molecules and receptor binding sites. Neuron 79, 308–321 (2013).
Puchner, E.M., Walter, J.M., Kasper, R., Huang, B. & Lim, W.A. Counting molecules in single organelles with superresolution microscopy allows tracking of the endosome maturation trajectory. Proc. Natl. Acad. Sci. USA 110, 16015–16020 (2013).
Schücker, K., Holm, T., Franke, C., Sauer, M. & Benavente, R. Elucidation of synaptonemal complex organization by super-resolution imaging with isotropic resolution. Proc. Natl. Acad. Sci. USA 112, 2029–2033 (2015).
Lampe, A., Haucke, V., Sigrist, S.J., Heilemann, M. & Schmoranzer, J. Multi-colour direct STORM with red emitting carbocyanines. Biol. Cell 104, 229–237 (2012).
Lehmann, M., Lichtner, G., Klenz, H. & Schmoranzer, J. Novel organic dyes for multicolor localization-based super-resolution microscopy. J. Biophotonics http://dx.doi.org/10.1002/jbio.201500119 (13 May 2015).
Huang, B., Wang, W., Bates, M. & Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008).
Lakadamyali, M., Babcock, H., Bates, M., Zhuang, X. & Lichtman, J. 3D multicolor super-resolution imaging offers improved accuracy in neuron tracing. PLoS ONE 7, e30826 (2012).
Min, J. et al. 3D high-density localization microscopy using hybrid astigmatic/ biplane imaging and sparse image reconstruction. Biomed. Opt. Express 5, 3935–3948 (2014).
Kim, D. et al. Correlative stochastic optical reconstruction microscopy and electron microscopy. PLoS ONE 10, e0124581 (2015).
Crossman, D.J., Hou, Y., Jayasinghe, I., Baddeley, D. & Soeller, C. Combining confocal and single molecule localisation microscopy: a correlative approach to multi-scale tissue imaging. Methods 88, 98–108 (2015).
Hou, Y. et al. Super-resolution fluorescence imaging to study cardiac biophysics: α-actinin distribution and Z-disk topologies in optically thick cardiac tissue slices. Prog. Biophys. Mol. Biol. 115, 328–339 (2014).
Suleiman, H. et al. Nanoscale protein architecture of the kidney glomerular basement membrane. Elife 2, e01149 (2013).
Bates, M., Dempsey, G.T., Chen, K.H. & Zhuang, X. Multicolor super-resolution fluorescence imaging via multi-parameter fluorophore detection. Chemphyschem 13, 99–107 (2012).
Babcock, H., Sigal, Y.M. & Zhuang, X. A high-density 3D localization algorithm for stochastic optical reconstruction microscopy. Opt. Nanoscopy 1, 6 (2012).
Holden, S.J., Uphoff, S. & Kapanidis, A.N. DAOSTORM: an algorithm for high-density super-resolution microscopy. Nat. Methods 8, 279–280 (2011).
Dedecker, P., Duwé, S., Neely, R.K. & Zhang, J. Localizer: fast, accurate, open-source, and modular software package for superresolution microscopy. J. Biomed. Opt. 17, 126008 (2012).
Hedde, P.N., Fuchs, J., Oswald, F., Wiedenmann, J. & Nienhaus, G.U. Online image analysis software for photoactivation localization microscopy. Nat. Methods 6, 689–690 (2009).
Henriques, R. et al. QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nat. Methods 7, 339–340 (2010).
Wolter, S. et al. rapidSTORM: accurate, fast open-source software for localization microscopy. Nat. Methods 9, 1040–1041 (2012).
MacGillavry, H.D., Song, Y., Raghavachari, S. & Blanpied, T.A. Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron 78, 615–622 (2013).
Ehmann, N. et al. Quantitative super-resolution imaging of Bruchpilot distinguishes active zone states. Nat. Commun. 5, 4650 (2014).
Lu, H.E., MacGillavry, H.D., Frost, N.A. & Blanpied, T.A. Multiple spatial and kinetic subpopulations of CaMKII in spines and dendrites as resolved by single-molecule tracking PALM. J. Neurosci. 34, 7600–7610 (2014).
Nair, D. et al. Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. J. Neurosci. 33, 13204–13224 (2013).
Márquez-Neila, P., Baumela, L. & Alvarez, L. A morphological approach to curvature-based evolution of curves and surfaces. IEEE Trans. Pattern Anal. Mach. Intell. 36, 2–17 (2014).
Lorincz, A. & Nusser, Z. Specificity of immunoreactions: the importance of testing specificity in each method. J. Neurosci. 28, 9083–9086 (2008).
Andersen, P., Bliss, T.V. & Skrede, K.K. Lamellar organization of hippocampal pathways. Exp. Brain Res. 13, 222–238 (1971).
Teyler, T.J. Brain slice preparation: hippocampus. Brain Res. Bull. 5, 391–403 (1980).
Song, I. & Semyanov, A. Single cell electrophysiologic recordings in hippocampal slices. Protoc. Exchange http://dx.doi.org/10.1038/protex.2011.253 (2011).
Thévenaz, P., Ruttimann, U.E. & Unser, M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7, 27–41 (1998).
Lee, S.H., Földy, C. & Soltesz, I. Distinct endocannabinoid control of GABA release at perisomatic and dendritic synapses in the hippocampus. J. Neurosci. 30, 7993–8000 (2010).
Huang, B., Jones, S.A., Brandenburg, B. & Zhuang, X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat. Methods 5, 1047–1052 (2008).
Thompson, R.E., Larson, D.R. & Webb, W.W. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 82, 2775–2783 (2002).
Shroff, H., Galbraith, C.G., Galbraith, J.A. & Betzig, E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods 5, 417–423 (2008).
Acknowledgements
The authors are grateful to B. Pintér for technical assistance and help with establishing the tissue handling protocol; E. Tischler for technical assistance; and E. Szabadits and R. Nyilas (Institute of Experimental Medicine, Hungarian Academy of Sciences) for help with optimizing the immunostaining. We also thank M. Ledri (Institute of Experimental Medicine, Hungarian Academy of Sciences) for providing brain samples, I. Kacskovics (Immunogenes, Ltd.) and G. Balla (Institute of Experimental Medicine, Hungarian Academy of Sciences) for providing antibodies, and A. Zimmer (Institute of Molecular Psychiatry, University of Bonn) for providing knockout mice. We are grateful to S. Turbucz for his help with coding the software. We acknowledge the help of the Nikon Microscopy Center at the Institute of Experimental Medicine, Nikon Europe, Nikon Austria and Auro-Science Consulting for kindly providing microscopy support. The authors are indebted to I. Sase and I. Burda from Nikon for their invaluable help with the application of N-STORM. This work was supported by the Hungarian Academy of Sciences Momentum Program LP-54/2013, by the European Research Council Starting grant no. 243153 and by the Wellcome Trust International Senior Research Fellowship (to I.K.).
Author information
Authors and Affiliations
Contributions
L.B. and B.D. designed and performed STORM experiments, developed the protocol and analysis tools, prepared the figures and wrote the manuscript. V.M. and A.H. developed analysis and visualization tools and coded the software. Z.I.L. prepared tissue samples and performed immunostaining. I.K. conceived the method, supervised the project and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Overlay of fluorescence images with STORM.
Brain sections containing biocytin-labeled neurons were immunostained against CB1 receptors, imaged in dSTORM mode, loaded into VividSTORM, and density filtered according to steps 1-45, following national and institutional regulations. The widefield fluorescence (160 nm pixel size, recorded on the EMCCD camera) (a), the confocal (80 nm pixel size) (b), and the deconvolved confocal (c) images of the same area of the sample are compared with each other. (d-f) The corresponding STORM image of the same area is also shown. Note that while the overlaid of the pixel-based fluorescence and the STORM imaging modalities is possible with each approach, the widefield image suffers from low resolution, as indicated by the blurred contour of objects.
Supplementary Figure 2 Controlling background in STORM imaging.
(a) Immunostaining against CB1 receptor; and dSTORM imaging and analysis were performed on mouse brain sections following national and institutional regulations according to steps 1-45. The specific labeling of the immunostaining appears on axon terminals decorated with CB1. (b) In sections obtained from a littermate CB1 knockout mouse, only a scattered background labeling could be observed. This experiment serves as the negative control for the specificity of the primary antibody. (c) As negative control experiment to establish the specificity of the secondary antibody and the general immunostaining process, brain sections from wild-type mice were treated following the same protocol, but omitting the CB1 primary antibody from the immunostaining. Note that only very few non-specific localization points remained on the sections. These control experiments are important, because of the single-molecule sensitivity of STORM imaging. The background labeling of extremely low intensity is also detected as non-specific localization points, which are indistinguishable from the localization points representing the specific signal. (d-f) The same regions are shown after density filtering applied in VividSTORM. Note that the scattered low density labeling is reduced, whereas the specific high density labeling is preserved. (g) The number of localization points is plotted for each condition from 3 images per group, and demonstrates an excellent signal-to-noise ratio. This graph also illustrates that both the primary and secondary antibodies can contribute to the background to a small extent. (h) The mean lateral localization accuracy is similar in all groups, indicating that signal and background localization points cannot be distinguished in the image
Supplementary Figure 3 Tissue processing for STORM microscopy.
(a) Embedding the fixed tissue samples (e.g. 300 µm-thick brain slices) into 2% agarose (step 12). (b) Trimming the block for cutting 20 µm-thick brain slices by a vibratome (step 15). (c) A key step after finishing the immunostaining performed on free-floating brain sections is the transfer of the sections to acetone-cleaned coverslips. It is critical to gradually remove the buffer under a stereoscope, while preventing the section from curling up (step 23). (d) A hippocampal section mounted and dried on coverslip. The sections are stored at 4°C in this form until imaging.
Supplementary Figure 4 Image registration using fiducial markers in VividSTORM.
The sample, containing labeled axon terminals of neurons and anti-CB1 immunostaining, was prepared as in Figure 1, according to steps 1-23, in accordance with national and institutional regulations. Before mounting, the sections were labeled with FluoSpheres (see Supplementary Methods), and then imaging and image processing was carried out following steps 24-35. (a) The corresponding confocal (with registration already applied from TurboReg according to step 34) and STORM images are loaded in VividSTORM. Note the lack of alignment between the corresponding confocal and STORM CB1 channels, which is the cumulative consequence of calibration inaccuracies, sample drift and other microscope instabilities. (b) Only the bead channels are displayed, revealing the position of the fiducial markers recorded on both cameras. Landmarks (arrows) are assigned using the manual selection in the VividSTORM registration tool, and are shown enlarged in inserts. (c) Affine transformation is performed on all channels of the confocal image based on the landmarks set up in (b), resulting in good alignment between the corresponding CB1 channels in both modalities across the field of view. The boxed axon terminal is shown enlarged on (d).
Supplementary Figure 5 Evaluating STORM image quality.
(a) Brain sections were taken from 3 mice following national and institutional regulations, and CB1 immunostaining, dSTORM imaging and analysis were performed according to steps 1-45. Images (n = 271) of labeled axon terminals were taken at various distances from the tissue surface, with equal illumination, camera and analysis parameters. The mean lateral localization accuracy per image is plotted against depth. Lateral localization accuracy decreased by 0.18 nm per micron, and a weak correlation was evident (Pearson’s p < 0.001). Thus, the nanoscale resolution of STORM images remains excellent in the upper 10 micron of the tissue section regardless of the depth of the investigated target profile. (b) To demonstrate the effect of imaging laser power density on the number and quality of blinking events, 3 images per condition were taken at equal depth and with equal settings, but with different laser intensities (at 647 nm). Note the reduced number of localization points at lower intensities. (c) The localization accuracy also diminished with lowered laser intensity. (d) To demonstrate the effect of different activator laser power levels, sections were immunostained against CB1 (crosses) or homer (circles) using activator-reporter dye pairs, and imaged and analyzed according to steps 1-45. Images (one per condition) were taken at equal depths and with equal settings, but with different activator (405 nm) laser intensities. Localization accuracy was deteriorating with increasing the activator laser power, due to the increased background level of the image, and to the increased overlap of peaks. (e) However, the number of detected localizations did not increase with activation, as high initial density was accompanied by faster decay. For clarity, only 3 conditions from CB1 stainings are shown. Note that these effects are not prominent when using dSTORM due to the lower efficiency of activation. (f) The calculated z coordinates in a typical image are plotted as a histogram. Note the reduced probability of detection at increasing distances from the focal plane, and the asymmetrical shape of the histogram due to spherical aberration.
Supplementary information
Bleaching, blinking and activation of an immunostained brain sample at the start of STORM imaging
A previously unbleached area of a brain section after immunostaining by using secondary antibodies coupled with Alexa405/Alexa647 activator-reporter dye pairs is imaged with 30 ms exposure time, and with the cylindrical lens for astigmatic 3D STORM inserted. The video shows the raw EMCCD camera image (with LUT settings applied) in real-time. In the beginning, both the activator (405 nm) and imaging (647 nm) lasers are set to 0%. When the imaging laser is set to maximum power, the image is saturated at first, and then it bleaches in a few seconds. Individual spontaneous blinking events are clearly observable. Upon illumination with short pulses of low-power activator laser, the rate of activation is transiently increased. (AVI 4516 kb)
Iterative active contour selection of the confocal image of an axon terminal
To generate a visual demonstration of the iterative active contour evolution, the deconvolved confocal image of an axon terminal was magnified using bicubic resampling. The active contour selection tool from VividSTORM was run on the image, returning the position of the contour in each iteration. Note that in the actual program, the algorithm runs on the raw image data, and only the end result is displayed. (AVI 233 kb)
Rights and permissions
About this article
Cite this article
Barna, L., Dudok, B., Miczán, V. et al. Correlated confocal and super-resolution imaging by VividSTORM. Nat Protoc 11, 163–183 (2016). https://doi.org/10.1038/nprot.2016.002
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.002
This article is cited by
-
Unique features of the arterial blood–brain barrier
Fluids and Barriers of the CNS (2023)
-
Discrimination of normal and cancerous human skin tissues based on laser-induced spectral shift fluorescence microscopy
Scientific Reports (2022)
-
A density-based enrichment measure for assessing colocalization in single-molecule localization microscopy data
Nature Communications (2022)
-
Robust adaptive optics for localization microscopy deep in complex tissue
Nature Communications (2021)
-
The cardiac nanoenvironment: form and function at the nanoscale
Biophysical Reviews (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.