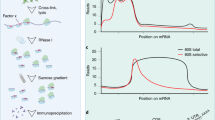
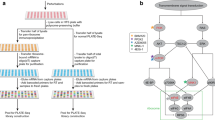
Abstract
Recent studies highlight the importance of translational control in determining protein abundance, underscoring the value of measuring gene expression at the level of translation. We present a protocol for genome-wide, quantitative analysis of in vivo translation by deep sequencing. This ribosome profiling approach maps the exact positions of ribosomes on transcripts by nuclease footprinting. The nuclease-protected mRNA fragments are converted into a DNA library suitable for deep sequencing using a strategy that minimizes bias. The abundance of different footprint fragments in deep sequencing data reports on the amount of translation of a gene. In addition, footprints reveal the exact regions of the transcriptome that are translated. To better define translated reading frames, we describe an adaptation that reveals the sites of translation initiation by pretreating cells with harringtonine to immobilize initiating ribosomes. The protocol we describe requires 5–7 days to generate a completed ribosome profiling sequencing library. Sequencing and data analysis require a further 4–5 days.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Change history
17 August 2012
In the version of this article initially published, the table in Step 32 of the protocol lists “T4 PNK (10 U µl–1)”, “1.0 µl” and “10 U” in the last row. This entry should read “T4 Rnl2(tr) (200 U µl–1)”, “1.0 µl” and “200 U”. The error has been corrected in the HTML and PDF versions of the article.
References
Brown, P.O. & Botstein, D. Exploring the new world of the genome with DNA microarrays. Nat. Genet. 21, 33–37 (1999).
Mortazavi, A., Williams, B.A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008).
Nagalakshmi, U. et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 (2008).
Ghaemmaghami, S. et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003).
Lu, P., Vogel, C., Wang, R., Yao, X. & Marcotte, E.M. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25, 117–124 (2007).
Schwanhausser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Vogel, C. Translation's coming of age. Mol. Syst. Biol. 7, 498 (2011).
Sonenberg, N. & Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 (2009).
Ingolia, N.T., Ghaemmaghami, S., Newman, J.R. & Weissman, J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
Steitz, J.A. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature 224, 957–964 (1969).
Wolin, S.L. & Walter, P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 7, 3559–3569 (1988).
Bentley, D.R. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008).
Ingolia, N.T., Lareau, L.F. & Weissman, J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011).
Hsieh, A.C. et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 (2012).
Kondo, T. et al. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329, 336–339 (2010).
Ingolia, N.T. Genome-wide translational profiling by ribosome footprinting. Methods Enzymol. 470, 119–142 (2010).
Brar, G.A. et al. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335, 552–557 (2011).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Dinger, M.E., Pang, K.C., Mercer, T.R. & Mattick, J.S. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 4, e1000176 (2008).
Robert, F. et al. Altering chemosensitivity by modulating translation elongation. PloS ONE 4, e5428 (2009).
Fresno, M., Jimenez, A. & Vazquez, D. Inhibition of translation in eukaryotic systems by harringtonine. Eur. J. Biochem. 72, 323–330 (1977).
Levin, J.Z. et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods 7, 709–715 (2010).
Li, G.W., Oh, E. & Weissman, J.S. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484, 538–541 (2012).
Qian, W., Yang, J.-R., Pearson, N.M., Maclean, C. & Zhang, J. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 8, e1002603 (2012).
Stadler, M. & Fire, A. Wobble base-pairing slows in vivo translation elongation in metazoans. RNA 17, 2063–2073 (2011).
Oh, E. et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147, 1295–1308 (2011).
Reid, D.W. & Nicchitta, C.V. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J. Biol. Chem. 287, 5518–5527 (2012).
Arava, Y. et al. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100, 3889–3894 (2003).
Sampath, P. et al. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2, 448–460 (2008).
del Prete, M.J., Vernal, R., Dolznig, H., Mullner, E.W. & Garcia-Sanz, J.A. Isolation of polysome-bound mRNA from solid tissues amenable for RT-PCR and profiling experiments. RNA 13, 414–421 (2007).
Blanchard, S.C., Kim, H.D., Gonzalez, R.L. Jr, Puglisi, J.D. & Chu, S. tRNA dynamics on the ribosome during translation. Proc. Natl. Acad. Sci. USA 101, 12893–12898 (2004).
Jelenc, P.C. Rapid purification of highly active ribosomes from Escherichia coli. Anal. Biochem. 105, 369–374 (1980).
Heiman, M. et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748 (2008).
Sanz, E. et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. USA 106, 13939–13944 (2009).
Lau, N.C., Lim, L.P., Weinstein, E.G. & Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862 (2001).
Hansen, K.D., Brenner, S.E. & Dudoit, S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 38, e131 (2010).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Bullard, J.H., Purdom, E., Hansen, K.D. & Dudoit, S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11, 94 (2010).
Li, J., Jiang, H. & Wong, W.H. Modeling non-uniformity in short-read rates in RNA-Seq data. Genome Biol. 11, R50 (2010).
Roberts, A., Trapnell, C., Donaghey, J., Rinn, J.L. & Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12, R22 (2011).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Hafner, M. et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA 17, 1697–1712 (2011).
Linsen, S.E. et al. Limitations and possibilities of small RNA digital gene expression profiling. Nat. Methods 6, 474–476 (2009).
Zhuang, F., Fuchs, R.T., Sun, Z., Zheng, Y. & Robb, G.B. Structural bias in T4 RNA ligase-mediated 3′-adapter ligation. Nucleic Acids Res. 40, e54 (2012).
Garber, M., Grabherr, M.G., Guttman, M. & Trapnell, C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods 8, 469–477 (2011).
Guo, H., Ingolia, N.T., Weissman, J.S. & Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 (2010).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Goecks, J., Nekrutenko, A. & Taylor, J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86 (2010).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Acknowledgements
We thank members of the Weissman and Ingolia labs, as well as H. Guo, D. Bartel, S. Luo and G. Schroth for advice in developing this protocol. This work was supported by the US National Institutes of Health (NIH) through an NIH P01 grant (AG10770; to J.S.W.) and a Ruth L. Kirschstein National Research Service Award (GM080853; to N.T.I.), an American Cancer Society postdoctoral fellowship (117945-PF-09-136-01-RMC; to G.A.B.) and the Searle Scholars Program (N.T.I.)
Author information
Authors and Affiliations
Contributions
N.T.I. and J.S.W. designed the study. G.A.B. and J.S.W. developed the rRNA depletion protocol. S.R. and J.S.W. adapted the protocol to use preadenylylated linker ligation. N.T.I., S.R., G.A.B. and A.M.M. performed experiments. N.T.I. and A.M.M. analyzed the data. N.T.I. and J.S.W. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
N.T.I. and J.S.W. are inventors on a patent covering the technique described in this manuscript.
Supplementary information
Supplementary Fig. 1
Comparison of expression measurements in different buffer conditions. The lower left-hand triangle in the comparison matrix compares per-gene expression measurements under two different buffer conditions, as in Fig. 2b. The upper right-hand triangle shows the histogram of log2 ratios in the comparison, as in Fig. 2b. (PDF 4635 kb)
Supplementary Fig. 2
Reading frame information in HEK293 samples prepared with different buffer conditions. (a-c) Stacked histograms showing the fraction of footprint reads at each length, separated based on the reading frame position of the 5′ end of the read, relative to the first codon nucleotide. (d-f) Histogram of footprint reads at each length, and of the information content of footprints at that length. The information content is defined as the difference between the entropy of the position distribution with no reading frame information, in which any of three codon positions are equally likely, and the entropy of the position distribution with reading frame information. (PDF 135 kb)
Supplementary Note
Galaxy workflow for ribosome footprinting analysis. This file contains a workflow that demonstrates the preprocessing and alignment of one million sequencing reads taken from the data presented here. This workflow requires the Galaxy software48. (ZIP 105251 kb)
Rights and permissions
About this article
Cite this article
Ingolia, N., Brar, G., Rouskin, S. et al. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7, 1534–1550 (2012). https://doi.org/10.1038/nprot.2012.086
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2012.086
This article is cited by
-
Ribosomal profiling of human endogenous retroviruses in healthy tissues
BMC Genomics (2024)
-
Ribosome profiling: a powerful tool in oncological research
Biomarker Research (2024)
-
Mesoscale DNA features impact APOBEC3A and APOBEC3B deaminase activity and shape tumor mutational landscapes
Nature Communications (2024)
-
Threonine fuels glioblastoma through YRDC-mediated codon-biased translational reprogramming
Nature Cancer (2024)
-
ChemRAP uncovers specific mRNA translation regulation via RNA 5′ phospho-methylation
EMBO Reports (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.