Abstract
The principal defining feature of Alzheimer's disease (AD) is memory impairment. As the transcription factor CREB (cAMP/Ca2+ responsive element-binding protein) is critical for memory formation across species, we investigated the role of CREB in a mouse model of AD. We found that TgCRND8 mice exhibit a profound impairment in the ability to form a spatial memory, a process that critically relies on the dorsal hippocampus. Perhaps contributing to this memory deficit, we observed additional deficits in the dorsal hippocampus of TgCRND8 mice in terms of (1) biochemistry (decreased CREB activation in the CA1 region), (2) neuronal structure (decreased spine density and dendritic complexity of CA1 pyramidal neurons), and (3) neuronal network activity (decreased arc mRNA levels following behavioral training). Locally and acutely increasing CREB function in the CA1 region of dorsal hippocampus of TgCRND8 mice was sufficient to restore function in each of these key domains (biochemistry, neuronal structure, network activity, and most importantly, memory formation). The rescue produced by increasing CREB was specific both anatomically and behaviorally and independent of plaque load or Aβ levels. Interestingly, humans with AD show poor spatial memory/navigation and AD brains have disrupted (1) CREB activation, and (2) spine density and dendritic complexity in hippocampal CA1 pyramidal neurons. These parallel findings not only confirm that TgCRND8 mice accurately model key aspects of human AD, but furthermore, suggest the intriguing possibility that targeting CREB may be a useful therapeutic strategy in treating humans with AD.
Similar content being viewed by others
INTRODUCTION
Alzheimer's disease (AD) is the most common form of dementia in the aging population. Although the precise molecular precursors of AD remain elusive, the majority of evidence indicates that β-amyloid (Aβ) has a key role in AD pathogenesis (Price et al, 1998; Mucke et al, 2000; Tanzi and Bertram, 2001; Hardy and Selkoe, 2002; Zhang et al, 2011). Aβ is formed by the sequential cleavage of amyloid precursor protein (APP) by β- and γ-secretases (Kang et al, 1987; Goate et al, 1991; Rossner et al, 1998; Selkoe, 1998; Mills and Reiner, 1999). Mutations in the genes encoding APP or presenilins (proteins that form part of the γ-secretase complex) increase Aβ levels (Citron et al, 1992, 1997; Cai et al, 1993; Suzuki et al, 1994) and are thought to cause familial forms of AD (Price et al, 1998; Hardy and Selkoe, 2002). Although increased levels of Aβ may eventually trigger cell death, memory impairment is often observed in AD patients before frank neurodegeneration (Cullen et al, 1997; Itoh et al, 1999; Selkoe, 2002; Vitolo et al, 2002). These findings suggest that high levels of Aβ negatively impact the molecular mechanisms required for normal memory formation.
CREB (cAMP/Ca2+ responsive element-binding protein) is a transcription factor that is critical for memory formation across a wide range of species (Dash et al, 1990; Bourtchuladze et al, 1994; Yin et al, 1995; Harum et al, 2001; Sekeres et al, 2010). Several distinct lines of evidence implicate mis-regulation of CREB function in AD. First, there is decreased CREB activation in post-mortem hippocampal tissue from AD patients (Yamamoto-Sasaki et al, 1999; Satoh et al, 2009). Second, analysis of an ‘AD transcriptome’ (produced from genome-wide expression profiling of CA1 hippocampal tissue from non-familial (sporadic) AD patients (Blalock et al, 2004)) identified CREB as the most highly connected hub in the AD transcriptome network (Satoh et al, 2009). Similar genomic network analysis shows a tight correlation between the number of connections (‘hubness’) of a gene and its ‘essentialness’ to a small-world network (Jeong et al, 2001), suggesting that disruption of CREB function is central to AD pathology. Third, exogenous application of oligomeric Aβ (Aβ1–42) blocks CREB function in cultured neurons, hippocampal slices and in vivo (Tong et al, 2001; Vitolo et al, 2002; Tong et al, 2004; Snyder et al, 2005; Garzon and Fahnestock, 2007; Spires-Jones et al, 2007). Similarly, expressing human genes encoding APP with familial AD mutations both increases Aβ levels and decreases CREB function in cultured cells (Ikezu et al, 1996; Arvanitis et al, 2007) and transgenic mice (Gong et al, 2004; Caccamo et al, 2011). Together, these diverse findings from both AD patients and various experimental preparations converge to suggest that the memory deficits observed in humans with AD may stem from decreased CREB function. Furthermore, these results suggest that directly increasing CREB function in brain regions important for memory may alleviate some of the memory deficits in AD patients.
Here, we directly tested whether increasing CREB function rescues the memory deficits observed in a transgenic mouse model of AD (TgCRND8 (Tg)). Importantly, these Tg mice show both decreased CREB function in the CA1 region of dorsal hippocampus, a region important for spatial memory, and profound deficits in the ability to form a spatial memory. We found that locally and acutely increasing CREB function specifically in the CA1 region of dorsal hippocampus was sufficient to rescue the spatial memory deficits of Tg mice, independent of changes in Aβ levels or plaque load. Together, these data show that disrupted CREB function in the CA1 region of dorsal hippocampus is a key regulator of the spatial memory impairment in a mouse model of AD and may have important implications for guiding the development of novel treatments for AD patients.
SUBJECTS AND METHODS
Mice
Tg mice express a human β APP695 transgene with both the Swedish (K670N-M671 L) and Indiana (V717F) familial AD mutations under the regulation of the Syrian hamster prion promoter (PrP) (Chishti et al, 2001) and were obtained from Dr David Westaway (University of Alberta, Canada). Tg mice were maintained in a 129S6/SvEvTac genetic background and crossed with wild-type (WT) C57BL/6NTac mice to produce F1 hybrid experimental mice. Tg and WT littermate control mice (an equal number of males and females) were 10 weeks old at the start of all experiments. For the αCaMKII experiment, we used adult mice with a deletion in exon 2 of αCaMKII (αCaMKIIΔ−/−), which results in a null mutation of αCaMKII protein (Elgersma et al, 2002). αCaMKIIΔ−/− and WT littermate controls were the F2 offspring of heterozygous mice (F1s were 50% C57BL/6NTac and 50% 129S6/SvEvTac). All mice were group housed (3–5 mice per cage) on a 12 h light–dark cycle. Behavioral experiments were conducted during the light phase. Food and water were available ad libitum. Procedures were approved by the Hospital for Sick Children Animal Care and Use Committee.
Immunocytochemistry
pCREB
To assess CREB function, we examined levels of pCREB (phosphorylated at Ser133) (Impey et al, 1998; Desmedt et al, 2003). Tg and WT mice were handled for 7 consecutive days and randomly assigned to the following behavioral treatment conditions; (1) homecage, (2) novel context exposure, and (3) watermaze probe test. Mice in the homecage condition were taken directly from their homecages. Mice in the novel context exploration condition were placed in a novel chamber and allowed to explore for 5 min, while mice in the watermaze probe condition were trained and tested in a watermaze (see below).
At 30 min following behavioral manipulation, mice were perfused transcardially with 4% paraformaldehyde. Brains were sliced coronally (50 μm) and prepared for immunocytochemistry using anti-pCREB rabbit polyclonal (06–519; 1 : 1000, Upstate Cell Signaling Solutions, NY) as a primary antibody and biotinylated goat anti-rabbit antibody (111-065-144; 1 : 1000, Jackson ImmunoResearch, West Grove, PA) as a secondary antibody. Staining was visualized using avidin–biotin peroxidase (Vector Laboratories, Burlingame, CA) coupled to diaminobenzidine (DAB) (Sigma-Aldrich, St Louis, MO). No staining was detected in the absence of primary or secondary antibodies. All sections were processed and imaged in parallel under identical conditions. The dorsal hippocampus was imaged using a microscope (Nikon Eclipse 80i) with a 10 × objective. Two experimenters unaware of the treatment condition or genotype used Image J software (NIH) to quantify the number of pCREB+ nuclei (Han et al, 2008). The total number of pCREB+ cells in the CA1 and CA3 regions of dorsal hippocampus (as defined by Paxinos and Franklin, 2001) was quantified bilaterally from at least six sections from comparable anteroposterior levels from each mouse. The density of pCREB+ nuclei (per 0.1 mm2 of tissue) was calculated per mouse and averaged for each group. We also examined levels of pCREB+ nuclei in the dentate gyrus (DG) (Supplementary Figures S1a and S1b) and levels of overall CREB (both phosphorylated and nonphosphorylated, tCREB) protein in the CA1 region (Supplementary Figures S1c and S1d).
Plaques
Coronal brain sections (50 μm) from Tg and WT littermate mice were treated with 1% hydrogen peroxide in 100% methanol (15 min), followed by 96% formic acid (10 min). Plaques were stained with mouse anti-Aβ-6E10 primary antibody (SIG-39320; 1 : 500, Covance, Emeryville, CA) followed by a biotinylated donkey anti-mouse secondary antibody (715-066-151; 1 : 500, Jackson ImmunoResearch) and visualized using avidin–biotin peroxidase coupled to DAB, as above. The number of plaques was manually counted in the dorsal hippocampus and frontal cortex by two observers unaware of the treatment condition. To examine total plaque volume (plaque burden) in the dorsal hippocampus and frontal cortex, the planar rotator probe of Stereo Investigator software (MBF) was used. Outlines of the dorsal hippocampus or frontal cortex were traced (over at least six brain sections per mouse) and the entire surface area was scanned using the ‘meander scan’ function to locate and measure plaque volume. To estimate plaque volume, the edges of each amyloid plaque were marked with the planar rotator probe. The total plaque volume was divided by the volume of the brain structure (dorsal hippocampus or frontal cortex) to calculate the percentage of volume occupied by plaques.
Western Blot for Total and Phosphorylated CREB Protein
At 30 min after exposure to a novel context, dorsal hippocampal tissue from Tg and WT mice was isolated and homogenized in cell lysis buffer (50 mM Tris, 0.25 M sucrose, 25 mM KCl, 5 mM MgCl2 with protease and phosphatase inhibitors). Protein concentrations of homogenates were determined and samples were resuspended in buffer (100 mM Tris-Cl, pH 6.8, 200 mM DTT, 4% SDS, 20% glycerol, and 0.2% bromophenol blue). Homogenates were separated by SDS-polyacrylamide gel electrophoresis and electroblotted onto PVDF transfer membranes for western blotting. Two separate gels examined pCREB and tCREB protein levels, respectively. Membranes were blocked for 1 h in 5% skimmed milk in TBS and incubated overnight at 4 °C with primary antibodies in 1% skimmed milk in TBS. Rabbit anti-pCREB (06-519; 1 : 1000; Upstate Cell Signaling, as above), which recognizes CREB protein that is phosphorylated at Ser133, and mouse anti-CREB (MAB5432; 1 : 2000, Chemicon International), which recognizes total CREB protein were used. An antibody against glyceraldehyde-3-phosphate (GAPDH) (MA 2118; 1 : 10 000, Cell Signaling Technologies, Danvers, MA) was used as an internal loading control for both blots. Following overnight incubation, membranes were washed with TBS, 0.1% Tween-20 for 10 min and incubated for 1 to 2 h at room temperature with an HRP-conjugated species-specific secondary antibody. Bands were visualized by exposure of the membranes to film following treatment with Enhanced Chemiluminescence Western Blotting Analysis System (GE Healthcare). The optical intensity of the bands associated with pCREB, tCREB was measured relative to their respective GAPDH bands on the same blot using Image J. Total pCREB density was averaged over GAPDH band intensity, in order to rule out variances in band density between blots.
Fluorescent In Situ Hybridization (FISH)
Cellular compartment analysis of temporal activity by fluorescent in situ hybridization (or catFISH) for arc mRNA was performed as described previously (Guzowski et al, 1999; Han et al, 2007). Brains were removed 5 min following the watermaze probe test or from homecage mice, rapidly dissected and stored at −80°C. Coronal sections (20 μm) were cut and thaw-mounted on slides, such that each slide contained sections from different treatment conditions (watermaze and homecage) and genotypes (Tg and WT). Digoxigenin-labeled arc antisense and sense riboprobes were generated. Arc riboprobes were hybridized with tissue overnight. The digoxigenin-labeled arc riboprobe was detected with anti-digoxigenin-HRP conjugate and visualized using a cyanine-5 (Cy5) substrate kit. Nuclei were counterstained with DAPI. Sections were viewed on a confocal microscope (Zeiss LSM710) and image stacks from the CA1 region of dorsal hippocampus were collected at 1 μm intervals using a 40 × objective. Laser and PMT settings were optimized for the detection of arc for each slide, but kept constant for all images acquired from the same slide. As the dynamics of arc transcription in the DG are different (Chawla et al, 2005; Ramirez-Amaya et al, 2005), we restricted our analysis to the CA1 region of dorsal hippocampus.
Manual cell counts were performed by two observers unaware of the treatment condition or genotype of the mouse. Small, bright uniformly DAPI-stained nuclei (<5 μm in diameter) from putative glial cells were not counted. All other whole nuclei cells (putative neurons) in the field of view within the CA1 region were counted. Two to four slides were analyzed for each mouse. The percentage of Arc+ neurons in the CA1 region (total number of Arc+/total number DAPI+ neurons) was calculated per mouse and averaged per group.
HSV Vectors
For these experiments, we used two CREB vectors. For the behavioral studies, we subcloned CREB cDNA into the HSV amplicon, HSV-PrpUC, as previously described (Han et al, 2007, 2008, 2009). To visualize transgene expression, GFP was fused to the 5′ end of CREB cDNA. Previous studies established that tagging CREB with GFP does not interfere with the functional activity of CREB (Chao et al, 2002). Transgene expression was regulated by the constitutive promoter for the HSV immediate-early gene IE 4/5. In this CREB vector, therefore, CREB and GFP are fused and GFP is predominantly localized to the nucleus. As a control, GFP cDNA was similarly subcloned into the HSV-PrpUC backbone.
To visualize dendritic architecture and spines, we constructed two additional viral vectors using the pHSV-p1005 amplicon backbone (Clark et al, 2002; Russo et al, 2009). In this bi-cistronic vector, CREB and GFP are not fused (CREB expression is driven by the HSV immediate-early gene IE4/5 as above and GFP expression is driven by a separate CMV promoter) (Supplementary Figure S8a). This allows infected cells to be filled by GFP. As a control for this vector, we used the pHSV-p1005 amplicon (that lacks CREB cDNA but expresses GFP under the same CMV promoter). Although the CMV promoter contains several CRE elements, we found no difference in GFP expression between p1005-CREB and p1005 amplicons (see Supplementary Figures S8b and S8c).
All virus was packaged using a replication-defective helper virus as previously described (Carlezon et al, 2000; Carlezon and Neve, 2003; Han et al, 2007, 2008, 2009) and purified on a sucrose gradient, pelleted and resuspended in 10% sucrose. The average titer of the recombinant virus stocks was typically 4.0 × 107 infectious units/ml. Previous studies indicate that microinjection of either CREB vector increased both CREB levels and CREB function (CRE-mediated transcription) (Barrot et al, 2002; Olson et al, 2005).
Surgery
Mice were pretreated with atropine sulfate (0.1 mg/kg, i.p.), anesthetized with chloral hydrate (400 mg/kg, i.p.) and placed in a stereotaxic frame. The skin was retracted and holes drilled in the skull bilaterally above the dorsal hippocampus (AP=−1.5, ML=±2.25, V=−1.75 mm from bregma) according to (Paxinos and Franklin, 2001). Bilateral microinjections of the vectors (2.0 μl) were delivered through glass micropipettes into the CA1 region over 20 min. Micropipettes were left in place an additional 10 min to ensure diffusion of the vector. As transgene expression using this viral system typically peaks 3 days following surgery (Barrot et al, 2002), we trained mice 1 day following surgery for 3 days in all experiments. We previously found that mice microinjected with GFP vector are able to form a robust spatial memory using this protocol (Sekeres et al, 2010).
Histology
An experimenter unaware of the behavioral data, vector microinjected or genotype of the mouse assessed placement and extent of viral infection for each mouse using GFP-fluorescence. Four days following vector microinjection, mice were perfused with 4% paraformaldehyde. Brains were sliced coronally (50 μm). Mice were classified as ‘hits’ and included in the subsequent data analysis if they had robust bilateral transgene expression in the CA1 region of dorsal hippocampus (see Supplementary Figure S7). Some mice showed no GFP expression in the CA1 region, but robust expression in the DG. The data from these mice were analyzed separately (Figures 3a–c). See Supplementary section for details of virus spread and quantification of number of neurons infected.
Watermaze Training and Spatial Memory Testing
The circular watermaze tank (120 cm diameter, 50 cm deep) was located in a dimly lit room (see Teixeira et al, 2006; Maei et al, 2009). The pool was filled to a depth of 40 cm with water made opaque by nontoxic white paint. Water temperature was maintained at 28±1 °C. A circular escape platform (10 cm diameter) was submerged 0.5 cm below the water surface and located in a fixed position throughout training. The pool was surrounded by white curtains painted with distinct cues, 1 m from the pool perimeter.
Before training, mice were handled for 2 min/day for 1 week. Mice received two blocks of three trials per day, for 3 days. Each trial lasted a maximum of 60 s. To begin each trial, mice were placed in the pool, facing the wall in one of four start locations (varied pseudo randomly). The trial was complete once the mouse found the platform or 60 s had elapsed. If the mouse failed to find the platform on any trial, the experimenter guided the mouse onto the platform. After each training trial, the mouse was allowed 15 s on top of the platform. At 60 min after the final training trial, spatial memory was assessed in a probe test during which the platform was removed from the pool and the mouse allowed to search for 60 s. It should be noted that using this robust training protocol we did not observe a memory enhancement in WT littermate mice microinjected with CREB vector, perhaps because these mice were already performing at ceiling levels.
Behavioral data from the training and probe trials were acquired and analyzed using an automated tracking system (Actimetrics, Wilmette, IL). For the training data, we analyzed the escape latency, swim speed, distance travelled, and thigmotaxis (the degree to which animals spend time in the perimeter of the pool) (see Supplementary section for details). In the probe test, our primary measure was the amount of time mice spent in the target zone (20 cm radius, centered on the location of the platform during training; 11% of the pool surface, see Figure 1a) vs the average time spent in the three other equivalently sized zones located in the other quadrants of the pool (Moser et al, 1993; Moser and Moser, 1998; de Hoz et al, 2004). The analysis of the time spent in the target zone has previously been shown to be a highly sensitive measure of spatial memory (Maei et al, 2009; Moser and Moser, 1998). In addition, we measured the swim speed, thigmotaxis, time in all four zones, and time in four quadrants (see Supplementary section for details).
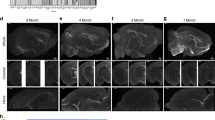
TgCRND8 mice show profound deficits in the ability to form a spatial memory and decreased CREB activity in the CA1 region of dorsal hippocampus. (a) Spatial memory in the probe test conducted after watermaze training was quantified by comparing the amount of time mice spent in the target (T) zone (20 cm radius, centered on the location of the platform during training; 11% of the pool surface) vs the average time spent in the three other (O) equivalently sized zones in the other quadrants areas of the pool (Maei et al, 2009). (b) Tg mice show deficits in the ability to form a spatial memory. WT littermate control mice (n=13) spent more time in the T zone compared with O zones of the pool, indicating the formation of spatial memory. However, Tg mice (n=9) did not form a spatial memory (mice searched T and O zones equally). Data in all graphs are mean±SEM. (c) Density plot for grouped data showing where WT and Tg mice concentrated their searches during the probe test. During training, the platform was located in the southeast zone of the pool (indicated by dotted circle). Color scale represents time spent per mouse per 5 × 5 cm2 area of the pool. As can be observed in this plot, WT mice concentrated their searches near the former platform location whereas Tg mice do not. (d) Tg mice have impaired CREB activation (pCREB) in the CA1 region of dorsal hippocampus following three different behavioral treatments. Photomicrographs of brains from WT and Tg mice either taken from the homecage (HC, top), exposed to a novel context (CX, middle) or trained and tested in the watermaze (WM, bottom). Low power examples of the entire dorsal hippocampus (left), and higher magnification images of the CA1 and CA3 regions of dorsal hippocampus (right). Images are quantified in (e) and (f). (e) The density of pCREB+ nuclei (per 100 μm2 tissue) from the CA1 region of dorsal hippocampus is lower in Tg mice than WT mice, across behavioral treatments (HC WT n=4, Tg n=4; CX WT n=7, Tg n=7; WM WT n=9, Tg n=9). (f) There is no difference in the density of pCREB+ nuclei in the CA3 region of dorsal hippocampus in Tg vs WT mice across behavioral treatments. (g, h) Western blot analysis confirms that Tg mice have lower pCREB levels in dorsal hippocampal than WT littermates following novel context exposure, but similar levels of total CREB (tCREB) levels. (g) Examples of immunoblots. Top band=pCREB (top example) or tCREB (bottom example), bottom band=GADPH (loading control). (h) Quantification of tCREB and pCREB protein levels from Tg (n=3) and WT (n=4) mice.
Statistical Analysis of Watermaze Data
To analyze the data from the watermaze probe test, we used an analysis of variance (ANOVA). First, we quantified spatial bias by comparing the time spent in the target zone vs the average time spent in equivalent and equally sized zones in the other three quadrants of the pool using an ANOVA with a between-subjects variable Vector and within-subjects variable Zone (Target, Others). This ANOVA asked whether each group searched selectively in the Target zone (acquired a spatial memory). Next, we directly compared the time spent in the target zone between groups using a one-way ANOVA. This analysis compared the degree of spatial memory across groups. Significant interactions or main effects were further analyzed using Newman–Keuls post hoc tests. The alpha value for all statistics held at 0.05.
Dendrite Complexity and Spine Analysis
Dendritic complexity and spine density of neurons infected by GFP or CREB (p1005) vectors were examined using GFP expression (see Supplementary Figure S8). We amplified the GFP signal using an antibody directed against GFP and DAB as a chromagen to visualize dendrites and dendritic spines using a light microscope. We chose this method because it prolongs the GFP signal, such that we could trace the entire cell and all its processes in the field of view (rather than dendritic segments) while avoiding potential photo-bleaching. Specifically, we used an anti-GFP primary rabbit polyclonal antibody (A11122; 1 : 1500, Invitrogen, Carlsbad, CA) and a biotinylated goat anti-rabbit secondary antibody (1 : 1000, Jackson ImmunoResearch). GFP+ pyramidal neurons in the CA1 region of dorsal hippocampus were imaged using a 100 × oil-immersion lens on a motorized microscope (Olympus BX61). First, we confirmed that our microinjections in the dorsal hippocampal CA1 region infected only pyramidal neurons by examining morphology. Infected cells had a triangular cell body located in the stratum pyramidale of the hippocampus with apical dendrites extending through the stratum radiatum, and stratum lacunosum-moleculare layer above the DG and basal dendrites extending into the stratum oriens, consistent with the characteristic morphology of a pyramidal neuron (Bannister and Larkman, 1995). The entire visible neuron (including cell body, apical and basal dendritic trees and all visible spines) was traced using Neurolucida software (MBF). Neurons were chosen for tracing based on their morphology (consistent with a pyramidal neuron), location (CA1 region of dorsal hippocampus), isolation (such that non-overlapping dendrites could be imaged), and general appearance (no obvious dendritic truncations).
Spines were defined according to Feldman and Dowd (1975) and included all protrusions that were in direct contact with the dendritic shaft. As this method has been shown to produce reliable results (Horner and Arbuthnott, 1991), we did not introduce a correction factor for hidden spines. For apical dendrites, dendritic spines at least 60 μm from the soma were counted and for basal dendrites, spines at least 20 μm from the soma were counted. Overall spine density was calculated per dendrite by dividing the total number of spines by the dendritic length and presented by averaging overall spine density per mouse on the apical and basal dendrites. The total number of spines, dendrites, and neurons counted for each group was as follows; WT-GFP (35 465 spines from 1051 dendrites from 14 neurons from n=7 mice) and WT-CREB (41 397 spines from 1291 dendrites from 12 neurons from n=6 mice), Tg-GFP (17 542 spines from 710 dendrites from 14 neurons from n=7 mice), and Tg-CREB (26 080 spines from 924 dendrites from 10 neurons from n=5 mice).
Statistical Analysis of Dendritic Architecture Data
To analyze dendritic complexity, we performed separate Group (WT-GFP, WT-CREB, Tg-GFP, and Tg-CREB) ANOVAs on (1) the number of intersections, (2) the dendritic length, and (3) the number of nodes, for basal and apical dendrites, respectively. To further evaluate significant differences, post hoc Newman–Keuls tests were used.
RESULTS
Tg Mice Show Profound Deficits in the Ability to Form a Spatial Memory
Allocentric spatial memory refers to the ability to represent spatial location independently of body orientation (O'Keefe and Nadel, 1978). AD patients have deficits in this type of spatial memory and navigation, even in the early stages of the disease (Henderson et al, 1989; deIpolyi et al, 2007; Jheng and Pai, 2009). To examine whether Tg mice are similarly impaired in an analogous form of spatial memory, we used the watermaze. In this task, mice learn to locate a platform hidden in a pool of opaque water using the distal cues around the pool for orientation, thereby mimicking real-world allocentric spatial memory and navigation (Ohta et al, 1981; Caplan and Lipman, 1995; Moffat et al, 2001).
We trained Tg and WT littermate control mice in the watermaze. At the end of training, we assessed whether mice had formed a spatial memory during a probe test during which the platform was removed from the pool (Figure 1a). During the probe test, WT mice spent more time in the area of the pool in which the platform was located during training (target zone (T), 20 cm radius circular zone centered on the former platform location) than in the other (O) equally sized areas of the pool, while Tg mice failed to selectively search in the target zone (Figures 1b and c). A Genotype (WT, Tg) × Zone (T, O) ANOVA revealed a significant interaction (F1,20=40.58; p<0.001), as well as significant main effects of Zone (F1,20=81.36; p<0.001) and Genotype (F1,20=27.68; p<0.001) alone. Post hoc comparisons showed that while WT mice searched selectively in the T zone, Tg mice showed no such preference. This interpretation was confirmed by an ANOVA specifically comparing time spent in T zone between Genotypes (F1,20=36.99; p<0.001). Therefore, Tg mice show profound deficits in the ability to form of a spatial memory.
This finding agrees with previous results using this (Chishti et al, 2001; Hyde et al, 2005) and other lines of transgenic mice developed to model human AD (including PDAPP mice (Chen et al, 2000), Tg2576 mice (Hsiao et al, 1996; Lesne et al, 2006), APP/PSEN1 bigenic mice (Arendash et al, 2001; Trinchese et al, 2004), and 3xTg-AD mice (Caccamo et al, 2011), especially female 3xTg-AD mice (Clinton et al, 2007)). Interestingly, human patients with AD and MCI (mild cognitive impairment, thought to be a prodrome of AD) (Petersen and Negash, 2008) also show deficits in humanized versions of the watermaze (Jheng and Pai, 2009; Laczo et al, 2010).
Tg mice have Decreased CREB Activation Specifically in the CA1 Region of Dorsal Hippocampus
Spatial memory in both humans (Feigenbaum and Morris, 2004; Parslow et al, 2005; Quiroga et al, 2005) and rodents (Jarrard, 1978; O'Keefe and Nadel, 1978; Nadel and McDonald, 1980; McNaughton et al, 1989) critically depends on the hippocampus. Furthermore, intact CREB function in the dorsal hippocampus, particularly in the CA1 region, is both necessary and sufficient for spatial memory formation in rodents (Guzowski and McGaugh, 1997; Pittenger et al, 2002; Sekeres et al, 2010). We hypothesized that disrupted CREB activity in this region could underlie the spatial memory deficits observed in the Tg mice. Therefore, we examined the levels of activated CREB (CREB that is phosphorylated at Ser133, pCREB) in Tg and WT littermate mice following three different behavioral treatments that induce varying levels of CREB activation in WT mice. Specifically, low pCREB levels are observed in WT mice taken directly from the homecage (Han et al, 2007), whereas higher levels are observed following novel context exploration (Vianna et al, 2000) or watermaze testing (Mizuno et al, 2002; Porte et al, 2008). We found that Tg mice had fewer pCREB+ neurons in the CA1 region of dorsal hippocampus than WT mice across all three behavioral treatments (Figures 1d and e). A Genotype × Treatment (homecage, novel context, watermaze) ANOVA revealed significant main effects of Genotype (F1,34=25.80; p<0.001) and Treatment (F2,34=136.79; p<0.001) but no Genotype × Treatment interaction (F2,34=1.96; p>0.05). Post hoc comparisons confirmed that Tg mice had fewer pCREB+ neurons in the CA1 region than WT mice across behavioral treatment.
The decrease in CREB activation in Tg mice was restricted to the CA1 region of dorsal hippocampus. There was no difference in pCREB levels in either the CA3 (Figure 1f; significant effect of Treatment (F2,34=97.75; p<0.001) only, no significant effect of Genotype × Treatment (F2,34=2.12; p>0.05) or Genotype (F1,34=3.92; p>0.05)) or dentate gyrus (DG) (Supplementary Figures S1a and S1b) regions of dorsal hippocampus.
We next asked whether Tg mice have lower overall levels of total CREB protein (both phosphorylated and non-phosphorylated CREB; tCREB). In contrast to the deficit in activated CREB, Tg mice showed normal levels of total CREB in the CA1 region. For instance, following novel context exploration we observed no decrease in tCREB levels in Tg mice using either immunohistochemistry (Figures S1c and S1d) or western blot analysis (Figures 1g and h; significant effects of Genotype × CREB (tCREB, pCREB) (F1,6=18.04; p<0.001), Genotype (F1,6=7.58; p<0.05), and CREB (F1,6=15.34; p<0.001). Post hoc comparisons confirmed that while tCREB levels did not differ, Tg mice had lower pCREB levels than WT mice). Interestingly, human brains from patients with AD also have disrupted CREB activation (Yamamoto-Sasaki et al, 1999; Satoh et al, 2009).
Locally and Acutely Increasing CREB Specifically in the CA1 Region of Dorsal Hippocampus Rescued the Spatial Memory Deficits in Tg Mice
We next examined whether increasing CREB function specifically in the CA1 region of dorsal hippocampus is sufficient to rescue the spatial memory deficits observed in Tg mice. We locally and acutely increased CREB function using replication-defective herpes simplex viral (HSV) vectors. We used HSV because, unlike many other viruses, HSV is naturally neurotropic (Fink et al, 1996). To increase CREB levels and function, we microinjected HSV encoding WT CREB (CREB vector). To visualize infected neurons, we fused CREB with GFP. HSV encoding GFP alone (GFP vector) served as a control. Tg and WT mice were microinjected with CREB or GFP vector 1 day before watermaze training. Vector microinjections produced robust bilateral transgene expression that was largely confined to the CA1 region of dorsal hippocampus (Figure 2a, Supplementary Figure S7).
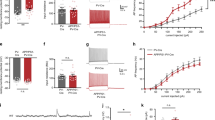
Acutely increasing CREB function in the CA1 region of dorsal hippocampus rescued the deficits in both spatial memory and hippocampal neuronal network activity in Tg mice. (a) Robust, localized transgene expression (GFP, green) following microinjection of CREB or GFP vectors into the CA1 region of dorsal hippocampus. Sections are counterstained with DAPI (blue). Low (right) and high (left) power examples of brains microinjected with GFP vector. (b) Microinjecting CREB vector into the CA1 region of dorsal hippocampus rescued the spatial memory deficit in Tg mice. Tg mice with GFP vector (Tg-GFP, n=14) show impaired spatial memory compared with WT littermates microinjected with GFP (n=16) or CREB (n=14) vector (WT mice spent more time in T zone compared with O zones of the pool (indicating the formation of spatial memory) whereas Tg-GFP mice did not search selectively in T zone). However, microinjecting CREB vector into the CA1 region of dorsal hippocampus rescued the memory deficit in Tg mice (Tg-CREB (n=11) mice spent equivalent time in T zone as WT mice). (c) Density plot for grouped data showing where WT and Tg mice microinjected with GFP or CREB vectors concentrated their searches during the probe test. The platform was located in the southeast area of the pool (dotted circle) during training. Color scale represents time spent per mouse per 5 × 5 cm2 area of the pool. As can be observed in this plot, WT mice (with GFP or CREB vector) concentrated their searches near the former platform location whereas Tg mice with GFP vector do not. Importantly, Tg mice microinjected with CREB vector performed similarly to WT mice. (d) In WT mice, training and testing in the watermaze increases the percent of recently active neurons in the CA1 region of dorsal hippocampus. WT mice trained and tested in the watermaze (WM) showed more Arc+ nuclei in the CA1 region of dorsal hippocampus than WT mice taken directly from the homecage (HC) (n=4 for each group). (e) Microinjecting CREB vector rescues the deficit in hippocampal network activity in Tg mice. Following the watermaze probe test, Tg mice with GFP vector (n=4) have fewer Arc+ nuclei in the CA1 region than similarly treated WT mice with GFP (n=7) or CREB (n=7) vector. Indeed, low level of Arc+ nuclei in Tg-GFP mice trained and tested in the watermaze was similar to WT mice taken directly from the homecage (d). However, Tg mice with CREB vector (n=7) showed equivalent Arc+ levels to WT mice trained and tested in the watermaze. The color reproduction of this figure is available at the Neuropsychopharmacology journal online.
Microinjecting CREB vector into the CA1 region of dorsal hippocampus not only restored normal CREB activity (Supplementary Figure S4) but also rescued the profound spatial memory deficits observed in Tg mice (Supplementary Figures 2b, 2c, and S2). Specifically, while Tg mice with GFP vector failed to search selectively in the target zone, Tg mice with CREB vector searched selectively in the target zone, showing a similar preference as their WT littermates (significant effects of Group × Zone (F3,51=4.59; p<0.001), Group (F3,51=8.59; p<0.001), and Zone (F1,34=64.95; p<0.001); post hoc comparisons confirmed that Tg-GFP mice showed no preference for T zone, but that Tg-CREB mice searched selectively in T zone. An ANOVA specifically comparing time spent in T zone between the groups supports this conclusion (F3,51=6.48; p<0.001). Post hoc comparisons verified that Tg-CREB mice and WT mice did not differ in time spent in T zone, while Tg-GFP mice spent significantly less time in T zone). Therefore, acutely restoring CREB function specifically in the CA1 region of dorsal hippocampus was sufficient to normalize spatial memory formation in Tg mice.
Acutely Increasing CREB Function in the CA1 Region of Dorsal Hippocampus of Tg Mice Rescued the Impairment in Hippocampal Neuronal Network Activity that may be Important for Spatial Memory
In agreement with the deficits in the ability to form a spatial memory, Tg mice also have disrupted activity in hippocampal neuronal networks that may be important for spatial memory. To investigate whether acutely increasing CREB levels and function also normalized this hippocampal neuronal network activity, we used catFISH to examine mRNA levels for the activity-dependent gene arc (activity-regulated cytoskeleton-associated protein; also known as Arg3.1). The similarities in the circuit dynamics between arc transcription and place cell firing (Guzowski et al, 1999; Vazdarjanova and Guzowski, 2004), suggest that this is a reliable method for assessing hippocampal cellular activity that reflects spatial information processing (Guzowski et al, 1999). As arc mRNA is localized to the nucleus 5–15 min following neuronal activity (Guzowski et al, 1999), we removed the brains of mice 5 min following the watermaze probe test. Neurons with arc signal localized to the nucleus (indicating neurons that were activated by the probe test) were designated as ‘Arc+’. Importantly, in WT mice, training and testing in the watermaze increases the percentage of Arc+ nuclei in the CA1 region of dorsal hippocampus (compared with WT mice taken directly from the homecage; Figure 2d; F1,30=141.99; p<0.001).
Following the watermaze probe test, Tg mice with GFP vector showed fewer Arc+ nuclei than WT mice (with either GFP or CREB vector) (Figure 2e). Indeed, Tg mice with GFP vector that were trained and tested in the watermaze showed similarly low levels of Arc+ nuclei to WT mice taken directly from the homecage (Figure 2d). However, microinjecting CREB vector restored arc levels in Tg mice to levels observed in trained WT mice (Figure 2e; F3,21=11.51; p<0.001, post hoc analysis confirms that only Tg-GFP mice show lower arc levels compared with WT mice (with either GFP or CREB vector)). Therefore, acutely increasing CREB levels in the CA1 region of dorsal hippocampus of Tg mice restored hippocampal neuronal network activity that may be important for spatial memory formation.
The Rescue of the Spatial Memory Deficit Produced by Increasing CREB in Tg Mice was Anatomically Specific
The rescue of the spatial memory deficit in Tg mice produced by microinjection of CREB vector was anatomically specific. Similar microinjection of CREB vector into the DG (Figure 3a) failed to reverse the impairment of spatial memory in Tg mice (Figures 3b and c, Supplementary Figure S3; Group (WT-DG-CREB, WT-CA1-CREB, Tg-DG-CREB, and Tg-CA1-CREB) × Zone ANOVA revealed a significant interaction (F3,36=7.98; p<0.001), as well as main effects of Group (F3,36=9.96; p<0.001) and Zone (F1,36=65.59; p<0.001). Post hoc comparisons showed that all groups, except Tg mice with CREB vector microinjected into DG, searched selectively in T zone. An ANOVA specifically comparing time spent in the T zone between groups confirms this interpretation (F3,36=10.82; p<0.001)). Therefore, acutely increasing CREB levels in the CA1 region of dorsal hippocampus, but not in DG, was sufficient to rescue the spatial memory deficit normally observed in this mouse model of AD.
The rescue of the spatial memory deficit in Tg mice by microinjecting the CREB vector was anatomically and behaviorally specific. (a) Example of vector microinjection (GFP, green) into the DG (rather than CA1 region) of dorsal hippocampus. Counterstained with DAPI (blue). (b) Microinjecting CREB vector into DG of Tg mice failed to rescue their spatial memory deficit (in contrast to the effects of microinjecting CREB vector into the CA1 region (Tg-CREB, from Figure 2b)). WT-CA1-CREB, n=14; WT-DG-CREB, n=5; Tg-CA1-CREB, n=11; Tg-DG-CREB, n=10. (c) Heat map for grouped data showing where Tg and WT mice microinjected with CREB vector into DG concentrated their searches during the probe trial. Color scale represents the number of visits per animal per 5 × 5 cm2 area of the pool. (d) Microinjecting CREB vector into the CA1 region of dorsal hippocampus does not rescue the spatial memory deficit of αCaMKIIΔ−/− mice. Both αCaMKIIΔ−/− mice with GFP (n=13) or CREB (n=13) vector show poor spatial memory, compared with WT littermates microinjected with GFP (n=5) or CREB (n=5) vector. (e) Density plot for grouped data showing where αCaMKIIΔ−/− and WT mice microinjected with GFP or CREB vectors concentrated their searches during the probe trial. Color scale represents the number of visits per animal per 5 × 5 cm2 area of the pool. The color reproduction of this figure is available at the Neuropsychopharmacology journal online.
The Rescue of the Spatial Memory Deficit Produced by Increasing CREB in the CA1 Region of Dorsal Hippocampus was Behaviorally Specific
To further examine the specificity of the rescue of spatial memory produced by microinjecting CREB vector into the CA1 region of Tg mice, we similarly microinjected CREB vector into another line of mice that also show profound spatial memory deficits. It is well-established that alpha-calcium calmodulin-dependent kinase II (αCaMKII) is critical for many forms of experience-dependent plasticity, including spatial memory formation (Silva et al, 1992; Lisman et al, 2002). αCaMKII may directly or indirectly phosphorylate CREB and is essential for L-type Ca2+ channel signaling to CREB (Takeda et al, 2007; Wheeler et al, 2008). We used mice homozygous for a deletion in αCaMKII Exon 2 (αCaMKIIΔ−/−, which results in a null mutation and the absence of αCaMKII protein (Elgersma et al, 2002)). Importantly, in these mice the mutation in αCaMKII is present throughout the brain, rather than being limited to the CA1 region of dorsal hippocampus. Therefore, we hypothesized that the spatial memory deficits observed in these αCaMKIIΔ−/− mutants would not be reversed by increasing CREB levels solely in the CA1 region of dorsal hippocampus.
Consistent with previous results (Elgersma et al, 2002), we found that αCaMKIIΔ−/− mice show spatial memory deficits in the watermaze (Figure 3d, Supplementary Figure S5). Importantly, microinjecting CREB vector into the CA1 region of dorsal hippocampus did not reverse this deficit (Figures 3d and e; Group × Zone (F3,32=4.41; p<0.05), Group (F3,32=10.22; p<0.001), and Zone (F1,32=16.47; p<0.001). Post hoc analyses revealed that αCaMKIIΔ−/− mice failed to show a preference for T zone, regardless of vector microinjected, while WT mice showed a preference for T zone, regardless of vector. An ANOVA specifically comparing time spent in the T zone between groups confirmed this interpretation (F3,32=8.44; p<0.001)). Therefore, increasing CREB levels in the CA1 region of dorsal hippocampus failed to restore the spatial memory deficits of mice with a brain-wide absence of αCaMKII protein. This finding contrasts the rescue produced by similar microinjection into Tg mice. Importantly, the disparate outcomes produced by microinjecting the same CREB vector indicate that increasing CREB function does not simply enhance nonspecific performance aspects of watermaze behavior (such as swimming, motivation to find the platform, and so on).
Increasing CREB Function in the CA1 Region of Dorsal Hippocampus did not Decrease Plaque Load in Tg Mice
To address the potential mechanisms by which CREB vector rescued the spatial memory deficit in Tg mice, we examined Aβ plaque load. Consistent with previous reports (Chishti et al, 2001; Hyde et al, 2005), we observed numerous Aβ plaques in dorsal hippocampus and frontal cortex of un-operated Tg mice, but no plaques in WT littermates (Figures 4a and b). Interestingly, microinjecting CREB vector into the CA1 region of dorsal hippocampus did not decrease the number of plaques in dorsal hippocampal (Figure 4c; F2,25=0.28; p>0.05) or frontal cortex (Figure 4d; F2,25=1.23; p>0.05) regions, compared with Tg mice without surgery. Similarly, the percent volume occupied by plaques in either dorsal hippocampus or frontal cortex of Tg mice was unaffected by CREB vector (Supplementary Figures S6a and S6b). This result is consistent with reports that Aβ plaque load does not correlate with cognitive function in people with AD (Katzman, 1988). In addition, we observed no difference in the high levels of aggregated or monomeric Aβ in the dorsal hippocampus of Tg mice microinjected with CREB vector compared with Tg mice microinjected with GFP vector or Tg mice without surgery (Supplementary Figures S6c and S6d). Together, these results indicate that increasing CREB function in dorsal hippocampus does not alleviate the memory deficits in Tg mice by decreasing plaque load or levels of aggregated Aβ.
The rescue of the spatial memory deficit in Tg mice by CREB was independent of Aβ plaque load. (a) Photomicrograph showing the presence of Aβ plaques in the dorsal hippocampus (HP) and frontal cortex (CO) regions of Tg mice. Low (left) and high (right) power representative images showing plaques in Tg mice. Importantly, age-matched WT littermate mice showed no Aβ plaques. (b) Plaques occupy roughly 6% of HP and CO volume in Tg mice (Tg mice without surgery, n=17). (c, d) Although microinjection of CREB vector rescued the spatial memory deficit in Tg mice, it did not reduce the number of plaques in either the dorsal hippocampus (c) or frontal cortex (d). Tg mice with GFP (n=5) or CREB (n=6) vectors show similar number of plaques as Tg mice that did not undergo surgery (n=17) in both the dorsal hippocampus and frontal cortex.
Increasing CREB Function Increased Dendritic Complexity in CA1 Pyramidal Neurons in Tg Mice
Post-mortem studies of human AD brains show striking changes in dendritic architecture in hippocampal CA1 pyramidal neurons (Yamada et al, 1988; el Hachimi and Foncin, 1990; Ferrer and Gullotta, 1990). Neurons from AD brains show both simplified dendritic architecture and decreased spine density suggesting that changes in neuronal morphology in this key brain region may cause the memory deficits of AD patients. Therefore, to examine whether Tg mice have an analogous deficit in neuronal morphology and whether CREB vector modulates this, we quantified dendritic complexity and spine density of CA1 pyramidal neurons infected by either CREB or GFP (p1005) vector (Figure 5a and b, Supplementary Figure S8).
Increasing CREB rescued the deficits in dendritic complexity and spine density in Tg dorsal hippocampus CA1 pyramidal neurons. (a) Representative image of immunostained pyramidal neurons from the CA1 region of dorsal hippocampus infected by GFP or CREB vector. (b) Sample tracing of infected pyramidal neuron showing apical and basal dendritic trees (generated by Neurolucida Explorer). (c, d) Tg neurons show decreased dendritic complexity along basal (c) dendrites (assessed by number of intersections (left), dendritic length (middle) and number of nodes (right)). This deficit in basal dendritic complexity in Tg mice is rescued by the CREB vector. Tg mice also show a deficit in dendritic length along the apical dendrites, a deficit that is reversed by CREB vector (d), (e) Representative basal dendritic segments of CA1 pyramidal neurons from Tg or WT mice infected with GFP or CREB vectors showing the distribution and density of dendritic spines. (f, g) Tg mice show decreased spine density (spines per μm dendrite) in the basal (f), but not apical (g), dendrites of CA1 pyramidal neurons compared with WT mice. (f) Microinjecting CREB vector normalized spine density along the basal dendrites of neurons from Tg mice to WT levels.
CA1 pyramidal neurons from Tg mice have simplified dendritic trees along the basal dendrites. Interestingly, CREB vector fully restored this (Figure 5c). Specifically, we observed fewer dendritic intersections in GFP-infected (control) neurons in Tg mice compared with WT mice (Tg-GFP vs WT-GFP neurons). CREB rescued this; the number of intersections in Tg-CREB neurons did not differ from WT-GFP neurons ((F3,46=19.49; p<0.001). Post hoc analyses confirmed that Tg mice had fewer dendritic intersections than WT mice, and that increasing CREB rescued this deficit in Tg neurons and further increased intersections in WT neurons).
Similar results were observed for dendritic length and the number of nodes along the basal dendrites of Tg CA1 pyramidal neurons. In particular, Tg mice had shorter basal dendrites that WT mice, a deficit that was reversed by CREB vector ((F3,46=23.93; p<0.001). Post hoc analyses showed that Tg-GFP dendrites were shorter than WT-GFP dendrites but that CREB restored length and further increased length in WT neurons). In addition, the decrease in nodes in Tg was also reversed by CREB vector ((F3,46=8.98; p<0.001). Post hoc analyses confirmed that Tg-GFP neurons had fewer nodes than WT-GFP neurons, but that the number of nodes was similar in Tg-CREB and WT-GFP neurons. CREB also increased nodes in WT mice). Therefore, using three measures of dendritic complexity, we observed that CA1 pyramidal neurons in Tg mice have simplified basal dendritic trees compared with WT mice. Importantly, CREB vector reversed this deficit in neuronal morphology in this critical brain region.
We observed more modest deficits in complexity along the apical dendrites of CA1 pyramidal neurons in Tg mice (Figure 5d). Although Tg-GFP neurons were not statistically different from WT-GFP neurons in terms of intersections or number of nodes, we did observe a significant decrease in dendritic length in Tg neurons compared with WT neurons. Specifically, an ANOVA conducted on the number of intersections showed an overall significant difference between groups (F3,46=4.62; p<0.001), but the crucial post hoc comparison revealed no statistical difference between the number of intersections in Tg-GFP compared with WT-GFP neurons (p>0.05). A similar pattern was observed for nodes (although the overall difference between groups was significant (F3,46=13.14; p<0.001), post hoc revealed no difference between Tg-GFP and WT-GFP neurons (p>0.05)). Interestingly, we observed a statistically significant decrease in the length of apical dendrites in Tg-GFP vs WT-GFP neurons, and this effect was reversed by CREB vector ((F3,46=14.00; p<0.001), post hoc analyses showed that Tg-GFP neurons were shorter than WT-GFP neurons, but that this was reversed by CREB vector and CREB also increased the length of WT neurons). Together, these data show that Tg mice have deficits in dendritic complexity, especially along the basal dendrites of CA1 pyramidal neurons, and that increasing CREB function restored normal morphology. Interestingly, using a similar analysis, human post-mortem AD brains also show a simplified dendritic tree along the basal dendrites of hippocampal CA1 neurons (Yamada et al, 1988). Therefore, acutely increasing CREB function rescued a validated deficit in neuronal structure in a mouse model of AD.
Increasing CREB Function Restored Spine Density in CA1 Dorsal Hippocampal Neurons in Tg Mice
A decrease in synapse number, particularly in the CA1 region of the hippocampus (Scheff et al, 2007), may better predict cognitive dysfunction in people with AD than either amyloid plaque or neurofibrillary tangle load (Terry et al, 1991; Lassmann et al, 1993; Coleman and Yao, 2003). As the majority of excitatory synapses on hippocampal CA1 pyramidal neurons occur on dendritic spines (Megias et al, 2001), we examined spine density in CA1 pyramidal neurons in Tg mice (Figures 5e–g). Tg mice showed decreased spine density on basal dendrites of CA1 pyramidal neurons (Tg-GFP neurons had a 46% reduction in spine density compared with WT-GFP neurons). This deficit was rescued by CREB vector (Figure 5e; F3,46=14.82; p<0.001, post hoc tests indicate that Tg-GFP neurons have fewer spines than WT-GFP neurons, but that CREB vector reverses this effect, while having no effect on WT neurons). As spine morphology may reflect function (Nimchinsky et al, 2002), we categorized spines into ‘mushroom’ and ‘non-mushroom’ classes. We observed a similar pattern of results for both mushroom and non-mushroom spines (data not shown). In contrast to the decrease in spine density along basal dendrites, Tg mice showed normal spine density along apical dendrites of CA1 neurons compared with WT mice (Figure 5g). Furthermore, increasing CREB had no effect on spine density on the apical dendrites of Tg mice (F3,46=2.59; p>0.05). In agreement with these findings, recent results from two additional mouse models of AD (Tg2576 and APP/Lo) showed decreased spine density along the basal (but not apical) dendrites of CA1 pyramidal neurons (Perez-Cruz et al, 2011).
DISCUSSION
The core feature of AD is an impairment in memory. Previous studies clearly established that the transcription factor CREB is critical for normal memory formation in species ranging from Aplysia and Drosophila to rats and mice (Bourtchuladze et al, 1994; Yin et al, 1995; Guzowski and McGaugh, 1997; Bartsch et al, 1998). Therefore, we examined the role of CREB in the memory deficits observed in a mouse model of AD. We used Tg mice and focused on the CA1 region of dorsal hippocampus because this region is among the first to exhibit pathology in humans with AD. We found that Tg mice exhibit a profound impairment in the ability to form a spatial memory, a task that critically relies on the dorsal hippocampus. Perhaps contributing to this memory deficit, we also observed deficits across multiple domains in the dorsal hippocampus of Tg mice, including (1) biochemistry (CREB activation), (2) neuronal structure (spine density and dendritic complexity of hippocampal CA1 pyramidal neurons), and (3) hippocampal neuronal network activity (arc mRNA levels following behavioral training). Moreover, locally and acutely increasing CREB function in the CA1 region of dorsal hippocampus of Tg mice was sufficient to restore function in each of these key domains (biochemistry, neuronal structure, network activity, and most importantly, memory formation). This rescue suggests that a disruption of dorsal hippocampal CREB function is at the heart of the memory deficits in this mouse model of AD. Interestingly, humans with AD also have poor spatial memory and navigation (Henderson et al, 1989; Pai and Jacobs, 2004; deIpolyi et al, 2007) and AD brains also have disrupted (1) CREB activation (Yamamoto-Sasaki et al, 1999; Satoh et al, 2009), and (2) spine density and dendritic complexity in hippocampal CA1 pyramidal neurons (Yamada et al, 1988; el Hachimi and Foncin, 1990; Ferrer and Gullotta, 1990). These parallel findings establish that Tg mice accurately model key aspects of human AD, and, furthermore, suggest that targeting CREB may be a useful therapeutic strategy in treating humans with AD. In these experiments, we assessed spatial memory 4 days following microinjection of CREB vector. To facilitate translation of these findings, it will be important to determine if more chronic increases in CREB function continue to produce similar beneficial effects on memory.
Tg mice showed deficits in both dorsal hippocampal CREB function and the ability to form a spatial memory. This is consistent with several previous findings that disrupting CREB function, specifically in the dorsal hippocampus, generally impairs the formation of spatial memory (Bourtchuladze et al, 1994; Guzowski and McGaugh, 1997; Pittenger et al, 2002). The spatial memory deficit observed in Tg mice was rescued by increasing CREB function in the CA1 region of dorsal hippocampus, but not DG. Importantly, Tg mice have intact CREB function in the DG. Alternatively, this anatomical specificity may reflect the different characteristics and function of these hippocampal subregions. Although both the CA1 and DG are critical for spatial memory formation (Jarrard, 1978; O'Keefe and Nadel, 1978; Nadel and McDonald, 1980; Castro et al, 1989; McNaughton et al, 1989; Kesner et al, 2002; Nakazawa et al, 2004; Bartsch et al, 2010), there are important differences. For instance, a smaller portion of DG neurons exhibit place fields (Jung and McNaughton, 1993; Leutgeb et al, 2007) or an increase in activity following spatial exploration (Chawla et al, 2005) or memory retrieval (Kee et al, 2007) compared with the CA1 region (O'Keefe, 1976; Guzowski et al, 1999). These data are consistent with theoretical notions that the DG sparsely encodes information (Marr, 1971; McNaughton and Morris, 1987; Rolls, 1990). Previous findings indicate that increasing CREB function increases neuronal excitability (Dong et al, 2006; Zhou et al, 2009). Therefore, it could be that microinjecting CREB vector into the DG did not rescue the spatial memory deficits of Tg mice because increasing neuronal excitability in the DG added ‘noise’ to the sparse code of the DG.
The rescue of the spatial memory deficits in Tg mice was also behaviorally specific. In contrast to the effects observed in Tg mice, increasing CA1 CREB function rescued failed to rescue the spatial memory deficits observed in αCaMKIIΔ−/− mutant mice. αCaMKIIΔ−/− mice have a deletion of αCaMKII that is brain-wide (not limited to the CA1 region of dorsal hippocampus). This lack of rescue indicates that increasing CREB in the dorsal hippocampus does not simply enhance the general motivation or motor behavior necessary for good performance in the watermaze task.
Memory formation is thought to be mediated by changes in synaptic efficacy (Bain, 1873; Bailey and Kandel, 1993; Bailey et al, 1996). As >90% of excitatory synapses in the hippocampus occur on dendritic spines (Harris and Stevens, 1989), the growth and re-structuring of spines may serve an important physical substrate underlying spatial memory formation. Indeed the loss of synapses may be the best correlate of memory impairment in humans with AD (DeKosky and Scheff, 1990; Terry et al, 1991). Similar to AD patients, we found that Tg mice have deficits in dendritic complexity and a decrease in spine density, especially along the basal dendrites of CA1 pyramidal neurons. Importantly, increasing CREB levels restored both deficits in Tg mice, suggesting that the restoration of normal neuronal morphology is critical for robust spatial memory formation.
The present finding that acutely increasing CREB function (only the order of days) increased dendritic complexity and spine density in Tg neurons is in agreement with previous results showing that CREB function is both necessary and sufficient for dendritic growth and arborization in hippocampal cultures (Wayman et al, 2006; Wayman et al, 2008). Moreover, increasing CREB function for only 24h has previously been shown to increase spine density in CA1 pyramidal neurons (Marie et al, 2005). It will be important to determine whether these changes in neuronal morphology are long lasting.
It is interesting to note that the observed deficits in dendritic complexity and spine density were greater in the basal dendrites of CA1 pyramidal neurons. Neuronal morphology along the apical dendrites was largely normal in Tg CA1 pyramidal neurons. There may be important anatomical and functional differences between basal and apical dendrites of CA1 pyramidal neurons. For instance, the basal dendrites preferentially receive projections from the contralateral CA3 region (Swanson et al, 1978; Ishizuka et al, 1990). In addition, a diverse array of treatments specifically increases spine density on basal CA1 dendrites. For instance, similar to the present finding, environmental enrichment (Moser et al, 1994, 1997), trace conditioning (Leuner et al, 2003) and context fear conditioning (Restivo et al, 2009) also increase spine density only on basal CA1 dendrites. These findings are consistent with the idea that basal dendritic synapses are more plastic than their apical counterparts (Kaibara and Leung, 1993) and, furthermore, suggest that a variety of manipulations increase spine density through a common mechanism, such as increasing CREB function.
Importantly, the present findings agree with, and extend, previous two key reports that three different treatments that may ultimately increase CREB function rescued the deficits observed in two AD mouse models. First, systemic administration of rolipram or UCh-L1 (ubiquitin carboxyl-terminal esterase L1) reversed the deficits in spine density (Smith et al, 2009) as well as a portion of the memory deficits (Gong et al, 2004; Gong et al, 2006) observed in APP/PS1 mice. Rolipram, a phosphodiesterase IV inhibitor, may increase CREB activation by preventing cAMP breakdown, which could lead to the activation of PKA and subsequent nuclear translocation of the catalytic subunits, where they may ultimately phosphorylate CREB. On the other hand, UCh-L1, a deubiquitinating enzyme, which removes ubiquitin from proteins that are destined to be metabolized via the proteasome pathway (Larsen et al, 1998), may promote the breakdown of PKA regulatory subunits, increasing the catalytic activity of PKA, and subsequently lead to CREB phosphorylation. However, both rolipram and Uch-L1 are not specific for CREB and may have additional ‘off-target’ effects, including interactions with microtubules, ligase activity and the degradation of additional substrates of the ubiquitin proteasome system (rather than specifically targeting the regulatory subunits of PKA) (Fimia and Sassone-Corsi, 2001; Liu et al, 2002; Seino and Shibasaki, 2005; Bheda et al, 2010; Boudreaux et al, 2010). Second, injection of lentivirus expressing CBP (CREB-binding protein) into the lateral ventricles of 6 month old 3xTg-AD mice rescued their spatial memory deficits (Caccamo et al, 2011). At this age, 3xTg-AD mice do not exhibit Aβ plaques, but do have high Aβ levels and a sexually dimorphic impairment in spatial memory formation (with females performing worse than males in the watermaze) (Clinton et al, 2007). Although named for its association with phosphorylated CREB (Chrivia et al, 1993), CBP is promiscuous and acts as a transcriptional co-activator for many DNA-binding proteins and also associates with dozens of nonhistone nuclear proteins (McManus and Hendzel, 2001; Hosoda et al, 2009). Therefore, although rolipram, Uch-L1 and CBP may ultimately enhance CREB function, none of these treatments are specific for CREB. Furthermore, systemic or lateral ventricular administration of these agents may not directly target the key brain regions important in spatial memory formation.
In this study, we found that directly increasing CREB levels specifically in the CA1 region of dorsal hippocampus produced a near complete rescue of the spatial memory deficits in Tg mice. Our findings that CREB vector also reversed the deficits in neuronal morphology and neuronal network activity in Tg mice suggest a mechanism mediating this rescue. The present results are in contrast to the partial restoration of memory previously reported following systemic treatment of rolipram or UCh-L1 (Gong et al, 2004, 2006). This suggests that the lack of complete rescue observed following rolipram or UCh-L1 may be due to ‘off target’ effects or the indirectness of these systemic treatments in terms molecules (CREB) or brain region (CA1 region of dorsal hippocampus). Nevertheless, all together, our current findings and previous reports show that the memory deficits observed in three different mouse models of AD were at least partially corrected using four diverse treatments that share a common property; these treatments increase CREB function. As a disruption of CREB function has been implicated in humans with AD (Yamamoto-Sasaki et al, 1999; Satoh et al, 2009), these findings converge to suggest that similarly targeting CREB may restore some cognitive function in human AD patients.
References
Arendash GW, King DL, Gordon MN, Morgan D, Hatcher JM, Hope CE et al (2001). Progressive, age-related behavioral impairments in transgenic mice carrying both mutant amyloid precursor protein and presenilin-1 transgenes. Brain Res 891: 42–53.
Arvanitis DN, Ducatenzeiler A, Ou JN, Grodstein E, Andrews SD, Tendulkar SR et al (2007). High intracellular concentrations of amyloid-beta block nuclear translocation of phosphorylated CREB. J Neurochem 103: 216–228.
Bailey CH, Bartsch D, Kandel ER (1996). Toward a molecular definition of long-term memory storage. Proc Natl Acad Sci USA 93: 13445–13452.
Bailey CH, Kandel ER (1993). Structural changes accompanying memory storage. Annu Rev Physiol 55: 397–426.
Bain A (1873). Mind and Body: The Theories of Their Relation. D. Appleton and Company: New York.
Bannister NJ, Larkman AU (1995). Dendritic morphology of CA1 pyramidal neurones from the rat hippocampus: II. Spine distributions. J Comp Neurol 360: 161–171.
Barrot M, Olivier JD, Perrotti LI, DiLeone RJ, Berton O, Eisch AJ et al (2002). CREB activity in the nucleus accumbens shell controls gating of behavioral responses to emotional stimuli. Proc Natl Acad Sci USA 99: 11435–11440.
Bartsch D, Casadio A, Karl KA, Serodio P, Kandel ER (1998). CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell 95: 211–223.
Bartsch T, Schonfeld R, Muller FJ, Alfke K, Leplow B, Aldenhoff J et al (2010). Focal lesions of human hippocampal CA1 neurons in transient global amnesia impair place memory. Science 328: 1412–1415.
Bheda A, Gullapalli A, Caplow M, Pagano JS, Shackelford J (2010). Ubiquitin editing enzyme UCH L1 and microtubule dynamics: implication in mitosis. Cell Cycle 9: 980–994.
Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW (2004). Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci USA 101: 2173–2178.
Boudreaux DA, Maiti TK, Davies CW, Das C (2010). Ubiquitin vinyl methyl ester binding orients the misaligned active site of the ubiquitin hydrolase UCHL1 into productive conformation. Proc Natl Acad Sci USA 107: 9117–9122.
Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva AJ (1994). Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 79: 59–68.
Caccamo A, Maldonado MA, Bokov AF, Majunder S, Oddo S (2011). CPB gene transfer increases BNDF levels and ameliorates learning and memory deficits in a mouse model of Alzheimer′s disease. Proc Natl Acad Sci USA 107: 22687–22692.
Cai XD, Golde TE, Younkin SG (1993). Release of excess amyloid beta protein from a mutant amyloid beta protein precursor. Science 259: 514–516.
Caplan LJ, Lipman PD (1995). Age and gender differences in the effectiveness of map-like learning aids in memory for routes. J Gerontol B Psychol Sci Soc Sci 50: P126–P133.
Carlezon Jr WA, Nestler EJ, Neve RL (2000). Herpes simplex virus-mediated gene transfer as a tool for neuropsychiatric research. Crit Rev Neurobiol 14: 47–67.
Carlezon Jr WA, Neve RL (2003). Viral-mediated gene transfer to study the behavioral correlates of CREB function in the nucleus accumbens of rats. Methods Mol Med 79: 331–350.
Castro CA, Silbert LH, McNaughton BL, Barnes CA (1989). Recovery of spatial learning deficits after decay of electrically induced synaptic enhancement in the hippocampus. Nature 342: 545–548.
Chao JR, Ni YG, Bolanos CA, Rahman Z, DiLeone RJ, Nestler EJ (2002). Characterization of the mouse adenylyl cyclase type VIII gene promoter: regulation by cAMP and CREB. Eur J Neurosci 16: 1284–1294.
Chawla MK, Guzowski JF, Ramirez-Amaya V, Lipa P, Hoffman KL, Marriott LK et al (2005). Sparse, environmentally selective expression of Arc RNA in the upper blade of the rodent fascia dentata by brief spatial experience. Hippocampus 15: 579–586.
Chen G, Chen KS, Knox J, Inglis J, Bernard A, Martin SJ et al (2000). A learning deficit related to age and beta-amyloid plaques in a mouse model of Alzheimer′s disease. Nature 408: 975–979.
Chishti MA, Yang DS, Janus C, Phinney AL, Horne P, Pearson J et al (2001). Early-onset amyloid deposition and cognitive deficits in transgenic mice expressing a double mutant form of amyloid precursor protein 695. J Biol Chem 276: 21562–21570.
Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH (1993). Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365: 855–859.
Citron M, Oltersdorf T, Haass C, McConlogue L, Hung AY, Seubert P et al (1992). Mutation of the beta-amyloid precursor protein in familial Alzheimer′s disease increases beta-protein production. Nature 360: 672–674.
Citron M, Westaway D, Xia W, Carlson G, Diehl T, Levesque G et al (1997). Mutant presenilins of Alzheimer′s disease increase production of 42-residue amyloid beta-protein in both transfected cells and transgenic mice. Nat Med 3: 67–72.
Clark MS, Sexton TJ, McClain M, Root D, Kohen R, Neumaier JF (2002). Overexpression of 5-HT1B receptor in dorsal raphe nucleus using Herpes Simplex Virus gene transfer increases anxiety behavior after inescapable stress. J Neurosci 22: 4550–4562.
Clinton LK, Billings LM, Green KN, Caccamo A, Ngo J, Oddo S et al (2007). Age-dependent sexual dimorphism in cognition and stress response in the 3xTg-AD mice. Neurobiol Dis 28: 76–82.
Coleman PD, Yao PJ (2003). Synaptic slaughter in Alzheimer′s disease. Neurobiol Aging 24: 1023–1027.
Cullen WK, Suh YH, Anwyl R, Rowan MJ (1997). Block of LTP in rat hippocampus in vivo by beta-amyloid precursor protein fragments. Neuroreport 8: 3213–3217.
Dash PK, Hochner B, Kandel ER (1990). Injection of the cAMP-responsive element into the nucleus of Aplysia sensory neurons blocks long-term facilitation. Nature 345: 718–721.
de Hoz L, Martin SJ, Morris RG (2004). Forgetting, reminding, and remembering: the retrieval of lost spatial memory. PLoS Biol 2: E225.
deIpolyi AR, Rankin KP, Mucke L, Miller BL, Gorno-Tempini ML (2007). Spatial cognition and the human navigation network in AD and MCI. Neurology 69: 986–997.
DeKosky ST, Scheff SW (1990). Synapse loss in frontal cortex biopsies in Alzheimer's disease: correlation with cognitive severity. Ann Neurol 27: 457–464.
Desmedt A, Hazvi S, Dudai Y (2003). Differential pattern of cAMP response element-binding protein activation in the rat brain after conditioned aversion as a function of the associative process engaged: taste versus context association. J Neurosci 23: 6102–6110.
Dong Y, Green T, Saal D, Marie H, Neve R, Nestler EJ et al (2006). CREB modulates excitability of nucleus accumbens neurons. Nat Neurosci 9: 475–477.
el Hachimi KH, Foncin JF (1990). Loss of dendritic spines in Alzheimer's disease. C R Acad Sci III 311: 397–402.
Elgersma Y, Fedorov NB, Ikonen S, Choi ES, Elgersma M, Carvalho OM et al (2002). Inhibitory autophosphorylation of CaMKII controls PSD association, plasticity, and learning. Neuron 36: 493–505.
Feigenbaum JD, Morris RG (2004). Allocentric versus egocentric spatial memory after unilateral temporal lobectomy in humans. Neuropsychology 18: 462–472.
Feldman ML, Dowd C (1975). Loss of dendritic spines in aging cerebral cortex. Anat Embryol (Berl) 148: 279–301.
Ferrer I, Gullotta F (1990). Down's syndrome and Alzheimer′s disease: dendritic spine counts in the hippocampus. Acta Neuropathol 79: 680–685.
Fimia GM, Sassone-Corsi P (2001). Cyclic AMP signalling. J Cell Sci 114: 1971–1972.
Fink DJ, DeLuca NA, Goins WF, Glorioso JC (1996). Gene transfer to neurons using herpes simplex virus-based vectors. Annu Rev Neurosci 19: 265–287.
Garzon DJ, Fahnestock M (2007). Oligomeric amyloid decreases basal levels of brain-derived neurotrophic factor (BDNF) mRNA via specific downregulation of BDNF transcripts IV and V in differentiated human neuroblastoma cells. J Neurosci 27: 2628–2635.
Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L et al (1991). Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer′s disease. Nature 349: 704–706.
Gong B, Cao Z, Zheng P, Vitolo OV, Liu S, Staniszewski A et al (2006). Ubiquitin hydrolase Uch-L1 rescues beta-amyloid-induced decreases in synaptic function and contextual memory. Cell 126: 775–788.
Gong B, Vitolo OV, Trinchese F, Liu S, Shelanski M, Arancio O (2004). Persistent improvement in synaptic and cognitive functions in an Alzheimer mouse model after rolipram treatment. J Clin Invest 114: 1624–1634.
Guzowski JF, McGaugh JL (1997). Antisense oligodeoxynucleotide-mediated disruption of hippocampal cAMP response element binding protein levels impairs consolidation of memory for water maze training. Proc Natl Acad Sci USA 94: 2693–2698.
Guzowski JF, McNaughton BL, Barnes CA, Worley PF (1999). Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci 2: 1120–1124.
Han JH, Kushner SA, Yiu AP, Cole CJ, Matynia A, Brown RA et al (2007). Neuronal competition and selection during memory formation. Science 316: 457–460.
Han JH, Kushner SA, Yiu AP, Hsiang HL, Buch T, Waisman A et al (2009). Selective erasure of a fear memory. Science 323: 1492–1496.
Han JH, Yiu AP, Cole CJ, Hsiang HL, Neve RL, Josselyn SA (2008). Increasing CREB in the auditory thalamus enhances memory and generalization of auditory conditioned fear. Learn Mem 15: 443–453.
Hardy J, Selkoe DJ (2002). The amyloid hypothesis of Alzheimer′s disease: progress and problems on the road to therapeutics. Science 297: 353–356.
Harris KM, Stevens JK (1989). Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: serial electron microscopy with reference to their biophysical characteristics. J Neurosci 9: 2982–2997.
Harum KH, Alemi L, Johnston MV (2001). Cognitive impairment in Coffin-Lowry syndrome correlates with reduced RSK2 activation. Neurology 56: 207–214.
Henderson VW, Mack W, Williams BW (1989). Spatial disorientation in Alzheimer′s disease. Arch Neurol 46: 391–394.
Horner CH, Arbuthnott E (1991). Methods of estimation of spine density--are spines evenly distributed throughout the dendritic field? J Anat 177: 179–184.
Hosoda H, Kato K, Asano H, Ito M, Kato H, Iwamoto T et al (2009). CBP/p300 is a cell type-specific modulator of CLOCK/BMAL1-mediated transcription. Mol Brain 2: 34.
Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S et al (1996). Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science 274: 99–102.
Hyde LA, Kazdoba TM, Grilli M, Lozza G, Brusa R, Zhang Q et al (2005). Age-progressing cognitive impairments and neuropathology in transgenic CRND8 mice. Behav Brain Res 160: 344–355.
Ikezu T, Okamoto T, Komatsuzaki K, Matsui T, Martyn JA, Nishimoto I (1996). Negative transactivation of cAMP response element by familial Alzheimer′s mutants of APP. EMBO J 15: 2468–2475.
Impey S, Smith DM, Obrietan K, Donahue R, Wade C, Storm DR (1998). Stimulation of cAMP response element (CRE)-mediated transcription during contextual learning. Nat Neurosci 1: 595–601.
Ishizuka N, Weber J, Amaral DG (1990). Organization of intrahippocampal projections originating from CA3 pyramidal cells in the rat. J Comp Neurol 295: 580–623.
Itoh A, Akaike T, Sokabe M, Nitta A, Iida R, Olariu A et al (1999). Impairments of long-term potentiation in hippocampal slices of beta-amyloid-infused rats. Eur J Pharmacol 382: 167–175.
Jarrard LE (1978). Selective hippocampal lesions: differential effects on performance by rats of a spatial task with preoperative versus postoperative training. J Comp Physiol Psychol 92: 1119–1127.
Jeong H, Mason SP, Barabasi AL, Oltvai ZN (2001). Lethality and centrality in protein networks. Nature 411: 41–42.
Jheng SS, Pai MC (2009). Cognitive map in patients with mild Alzheimer′s disease: a computer-generated arena study. Behav Brain Res 200: 42–47.
Jung MW, McNaughton BL (1993). Spatial selectivity of unit activity in the hippocampal granular layer. Hippocampus 3: 165–182.
Kaibara T, Leung LS (1993). Basal versus apical dendritic long-term potentiation of commissural afferents to hippocampal CA1: a current-source density study. J Neurosci 13: 2391–2404.
Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH et al (1987). The precursor of Alzheimer′s disease amyloid A4 protein resembles a cell-surface receptor. Nature 325: 733–736.
Katzman R (1988). Alzheimer's disease as an age-dependent disorder. Ciba Found Symp 134: 69–85.
Kee N, Teixeira CM, Wang AH, Frankland PW (2007). Preferential incorporation of adult-generated granule cells into spatial memory networks in the dentate gyrus. Nat Neurosci 10: 355–362.
Kesner RP, Lee I, Gilbert PE (2002). A behavioral assessment of hippocampal function based on subregional analysis. Rev Neurosci 15: 333–351.
Laczo J, Andel R, Vyhnalek M, Vlcek K, Magerova H, Varjassyova A et al (2010). Human analogue of the morris water maze for testing subjects at risk of Alzheimer's disease. Neurodegener Dis 7: 148–152.
Larsen CN, Krantz BA, Wilkinson KD (1998). Substrate specificity of deubiquitinating enzymes: ubiquitin C-terminal hydrolases. Biochemistry 37: 3358–3368.
Lassmann H, Fischer P, Jellinger K (1993). Synaptic pathology of Alzheimer's disease. Ann NY Acad Sci 695: 59–64.
Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A et al (2006). A specific amyloid-beta protein assembly in the brain impairs memory. Nature 440: 352–357.
Leuner B, Falduto J, Shors TJ (2003). Associative memory formation increases the observation of dendritic spines in the hippocampus. J Neurosci 23: 659–665.
Leutgeb JK, Leutgeb S, Moser MB, Moser EI (2007). Pattern separation in the dentate gyrus and CA3 of the hippocampus. Science 315: 961–966.
Lisman J, Schulman H, Cline H (2002). The molecular basis of CaMKII function in synaptic and behavioural memory. Nat Rev Neurosci 3: 175–190.
Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury Jr PT (2002). The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson′s disease susceptibility. Cell 111: 209–218.
Maei HR, Zaslavsky K, Teixeira CM, Frankland PW (2009). What is the most sensitive measure of water maze probe test performance? Front Integr Neurosci 3: 4.
Marie H, Morishita W, Yu X, Calakos N, Malenka RC (2005). Generation of silent synapses by acute in vivo expression of CaMKIV and CREB. Neuron 45: 741–752.
Marr A (1971). Simple memory: a theory for archicortex. Philos Trans R Soc Lond B Biol Sci 262: 23–81.
McManus KJ, Hendzel MJ (2001). CBP, a transcriptional coactivator and acetyltransferase. Biochem Cell Biol 79: 253–266.
McNaughton BL, Barnes CA, Meltzer J, Sutherland RJ (1989). Hippocampal granule cells are necessary for normal spatial learning but not for spatially-selective pyramidal cell discharge. Exp Brain Res 76: 485–496.
McNaughton BL, Morris RGM (1987). Hippocampal synpatic enhancement and information storage within a distributed memory systems. Trends Neurosci 10: 408–415.
Megias M, Emri Z, Freund TF, Gulyas AI (2001). Total number and distribution of inhibitory and excitatory synapses on hippocampal CA1 pyramidal cells. Neuroscience 102: 527–540.
Mills J, Reiner PB (1999). Regulation of amyloid precursor protein cleavage. J Neurochem 72: 443–460.
Mizuno M, Yamada K, Maekawa N, Saito K, Seishima M, Nabeshima T (2002). CREB phosphorylation as a molecular marker of memory processing in the hippocampus for spatial learning. Behav Brain Res 133: 135–141.
Moffat SD, Zonderman AB, Resnick SM (2001). Age differences in spatial memory in a virtual environment navigation task. Neurobiol Aging 22: 787–796.
Moser E, Moser MB, Andersen P (1993). Spatial learning impairment parallels the magnitude of dorsal hippocampal lesions, but is hardly present following ventral lesions. J Neurosci 13: 3916–3925.
Moser MB, Moser EI (1998). Distributed encoding and retrieval of spatial memory in the hippocampus. J Neurosci 18: 7535–7542.
Moser MB, Trommald M, Andersen P (1994). An increase in dendritic spine density on hippocampal CA1 pyramidal cells following spatial learning in adult rats suggests the formation of new synapses. Proc Natl Acad Sci USA 91: 12673–12675.
Moser MB, Trommald M, Egeland T, Andersen P (1997). Spatial training in a complex environment and isolation alter the spine distribution differently in rat CA1 pyramidal cells. J Comp Neurol 380: 373–381.
Mucke L, Masliah E, Yu GQ, Mallory M, Rockenstein EM, Tatsuno G et al (2000). High-level neuronal expression of abeta 1-42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J Neurosci 20: 4050–4058.
Nadel L, McDonald L (1980). Hippocampus: cognitive map or working memory? Behav Neurol Biol 29: 405–409.
Nakazawa K, McHugh TJ, Wilson MA, Tonegawa S (2004). NMDA receptors, place cells and hippocampal spatial memory. Nat Rev Neurosci 5: 361–372.
Nimchinsky EA, Sabatini BL, Svoboda K (2002). Structure and function of dendritic spines. Annu Rev Physiol 64: 313–353.
O'Keefe J (1976). Place units in the hippocampus of the freely moving rat. Exp Neurol 51: 78–109.
O'Keefe J, Nadel L (1978). The Hippocampus as a Cognitive Map. Clarendon Press: London.
Ohta RJ, Walsh DA, Krauss IK (1981). Spatial perspective-taking ability in young and elderly adults. Exp Aging Res 7: 45–63.
Olson VG, Zabetian CP, Bolanos CA, Edwards S, Barrot M, Eisch AJ et al (2005). Regulation of drug reward by cAMP response element-binding protein: evidence for two functionally distinct subregions of the ventral tegmental area. J Neurosci 25: 5553–5562.
Pai MC, Jacobs WJ (2004). Topographical disorientation in community-residing patients with Alzheimer′s disease. Int J Geriatr Psychiatry 19: 250–255.
Parslow DM, Morris RG, Fleminger S, Rahman Q, Abrahams S, Recce M (2005). Allocentric spatial memory in humans with hippocampal lesions. Acta Psychol (Amst) 118: 123–147.
Paxinos G, Franklin KBJ (2001). The Mouse Brain in Stereotaxic Coordinates, 2nd edn. Academic Press: San Diego.
Perez-Cruz C, Nolte MW, van Gaalen MM, Rustay NR, Termont A, Tanghe A et al (2011). Reduced spine density in specific regions of CA1 pyramidal neurons in two transgenic mouse models of Alzheimer′s disease. J Neurosci 31: 3926–3934.
Petersen RC, Negash S (2008). Mild cognitive impairment: an overview. CNS Spectr 13: 45–53.
Pittenger C, Huang YY, Paletzki RF, Bourtchouladze R, Scanlin H, Vronskaya S et al (2002). Reversible inhibition of CREB/ATF transcription factors in region CA1 of the dorsal hippocampus disrupts hippocampus-dependent spatial memory. Neuron 34: 447–462.
Porte Y, Buhot MC, Mons NE (2008). Spatial memory in the Morris water maze and activation of cyclic AMP response element-binding (CREB) protein within the mouse hippocampus. Learn Mem 15: 885–894.
Price DL, Tanzi RE, Borchelt DR, Sisodia SS (1998). Alzheimer′s disease: genetic studies and transgenic models. Annu Rev Genet 32: 461–493.
Quiroga RQ, Reddy L, Kreiman G, Koch C, Fried I (2005). Invariant visual representation by single neurons in the human brain. Nature 435: 1102–1107.
Ramirez-Amaya V, Vazdarjanova A, Mikhael D, Rosi S, Worley PF, Barnes CA (2005). Spatial exploration-induced Arc mRNA and protein expression: evidence for selective, network-specific reactivation. J Neurosci 25: 1761–1768.
Restivo L, Vetere G, Bontempi B, Ammassari-Teule M (2009). The formation of recent and remote memory is associated with time-dependent formation of dendritic spines in the hippocampus and anterior cingulate cortex. J Neurosci 29: 8206–8214.
Rolls ET (1990). Theoretical and neurophysiological analysis of the functions of the primate hippocampus in memory. Cold Spring Harb Symp Quant Biol 55: 995–1006.
Rossner S, Ueberham U, Schliebs R, Perez-Polo JR, Bigl V (1998). The regulation of amyloid precursor protein metabolism by cholinergic mechanisms and neurotrophin receptor signaling. Prog Neurobiol 56: 541–569.
Russo SJ, Wilkinson MB, Mazei-Robison MS, Dietz DM, Maze I, Krishnan V et al (2009). Nuclear factor kappa B signaling regulates neuronal morphology and cocaine reward. J Neurosci 29: 3529–3537.
Satoh J, Tabunoki H, Arima K (2009). Molecular network analysis suggests aberrant CREB-mediated gene regulation in the Alzheimer disease hippocampus. Dis Markers 27: 239–252.
Scheff SW, Price DA, Schmitt FA, DeKosky ST, Mufson EJ (2007). Synaptic alterations in CA1 in mild Alzheimer disease and mild cognitive impairment. Neurology 68: 1501–1508.
Seino S, Shibasaki T (2005). PKA-dependent and PKA-independent pathways for cAMP-regulated exocytosis. Physiol Rev 85: 1303–1342.
Sekeres MJ, Neve RL, Frankland PW, Josselyn SA (2010). Dorsal hippocampal CREB is both necessary and sufficient for spatial memory. Learn Mem 17: 280–283.
Selkoe DJ (1998). The cell biology of beta-amyloid precursor protein and presenilin in Alzheimer′s disease. Trends Cell Biol 8: 447–453.
Selkoe DJ (2002). Alzheimer's disease is a synaptic failure. Science 298: 789–791.
Silva AJ, Paylor R, Wehner JM, Tonegawa S (1992). Impaired spatial learning in alpha-calcium-calmodulin kinase II mutant mice. Science 257: 206–211.
Smith DL, Pozueta J, Gong B, Arancio O, Shelanski M (2009). Reversal of long-term dendritic spine alterations in Alzheimer disease models. Proc Natl Acad Sci USA 106: 16877–16882.
Snyder EM, Nong Y, Almeida CG, Paul S, Moran T, Choi EY et al (2005). Regulation of NMDA receptor trafficking by amyloid-beta. Nat Neurosci 8: 1051–1058.
Spires-Jones TL, Meyer-Luehmann M, Osetek JD, Jones PB, Stern EA, Bacskai BJ et al (2007). Impaired spine stability underlies plaque-related spine loss in an Alzheimer's disease mouse model. Am J Pathol 171: 1304–1311.
Suzuki N, Cheung TT, Cai XD, Odaka A, Otvos Jr L, Eckman C et al (1994). An increased percentage of long amyloid beta protein secreted by familial amyloid beta protein precursor (beta APP717) mutants. Science 264: 1336–1340.
Swanson LW, Wyss JM, Cowan WM (1978). An autoradiographic study of the organization of intrahippocampal association pathways in the rat. J Comp Neurol 181: 681–715.
Takeda H, Kitaoka Y, Hayashi Y, Kumai T, Munemasa Y, Fujino H et al (2007). Calcium/calmodulin-dependent protein kinase II regulates the phosphorylation of CREB in NMDA-induced retinal neurotoxicity. Brain Res 1184: 306–315.
Tanzi RE, Bertram L (2001). New frontiers in Alzheimer′s disease genetics. Neuron 32: 181–184.
Teixeira CM, Pomedli SR, Maei HR, Kee N, Frankland PW (2006). Involvement of the anterior cingulate cortex in the expression of remote spatial memory. J Neurosci 26: 7555–7564.
Terry RD, Masliah E, Salmon DP, Butters N, DeTeresa R, Hill R et al (1991). Physical basis of cognitive alterations in Alzheimer′s disease: synapse loss is the major correlate of cognitive impairment. Ann Neurol 30: 572–580.
Tong L, Balazs R, Thornton PL, Cotman CW (2004). Beta-amyloid peptide at sublethal concentrations downregulates brain-derived neurotrophic factor functions in cultured cortical neurons. J Neurosci 24: 6799–6809.
Tong L, Thornton PL, Balazs R, Cotman CW (2001). Beta-amyloid-(1-42) impairs activity-dependent cAMP-response element-binding protein signaling in neurons at concentrations in which cell survival is not compromised. J Biol Chem 276: 17301–17306.
Trinchese F, Liu S, Battaglia F, Walter S, Mathews PM, Arancio O (2004). Progressive age-related development of Alzheimer-like pathology in APP/PS1 mice. Ann Neurol 55: 801–814.
Vazdarjanova A, Guzowski JF (2004). Differences in hippocampal neuronal population responses to modifications of an environmental context: evidence for distinct, yet complementary, functions of CA3 and CA1 ensembles. J Neurosci 24: 6489–6496.
Vianna MR, Alonso M, Viola H, Quevedo J, de Paris F, Furman M et al (2000). Role of hippocampal signaling pathways in long-term memory formation of a nonassociative learning task in the rat. Learn Mem 7: 333–340.
Vitolo OV, Sant′Angelo A, Costanzo V, Battaglia F, Arancio O, Shelanski M (2002). Amyloid beta -peptide inhibition of the PKA/CREB pathway and long-term potentiation: reversibility by drugs that enhance cAMP signaling. Proc Natl Acad Sci USA 99: 13217–13221.
Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY et al (2008). An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci USA 105: 9093–9098.
Wayman GA, Impey S, Marks D, Saneyoshi T, Grant WF, Derkach V et al (2006). Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron 50: 897–909.
Wheeler DG, Barrett CF, Groth RD, Safa P, Tsien RW (2008). CaMKII locally encodes L-type channel activity to signal to nuclear CREB in excitation-transcription coupling. J Cell Biol 183: 849–863.
Yamada M, Wada Y, Tsukagoshi H, Otomo E, Hayakawa M (1988). A quantitative Golgi study of basal dendrites of hippocampal CA1 pyramidal cells in senile dementia of Alzheimer type. J Neurol Neurosurg Psychiatry 51: 1088–1090.
Yamamoto-Sasaki M, Ozawa H, Saito T, Rosler M, Riederer P (1999). Impaired phosphorylation of cyclic AMP response element binding protein in the hippocampus of dementia of the Alzheimer type. Brain Res 824: 300–303.
Yin JC, Del Vecchio M, Zhou H, Tully T (1995). CREB as a memory modulator: induced expression of a dCREB2 activator isoform enhances long-term memory in Drosophila. Cell 81: 107–115.
Zhang YW, Thompson R, Zhang H, Xu H (2011). APP processing in Alzheimer's disease. Mol Brain 4: 3.
Zhou Y, Won J, Karlsson MG, Zhou M, Rogerson T, Balaji J et al (2009). CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nat Neurosci 12: 1438–1443.
Acknowledgements
We thank Rachael Neve for packaging our initial virus, Valentina Mercaldo for technical expertise, Paul W Frankland and Leigh CP Botly for detailed comments on this paper and members of the Josselyn/Frankland lab for their helpful suggestions throughout this project. This work was supported by grants from the Canadian Institutes of Health Research (CIHR; MOP-74650), EJLB Foundation and the Alzheimer's Society of Canada. APY and AJR received support from Restracomp Fellowships (Hospital for Sick Children) and APY from the Alzheimer's Society of Canada.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Neuropsychopharmacology website
Supplementary information
Rights and permissions
About this article
Cite this article
Yiu, A., Rashid, A. & Josselyn, S. Increasing CREB Function in the CA1 Region of Dorsal Hippocampus Rescues the Spatial Memory Deficits in a Mouse Model of Alzheimer's Disease. Neuropsychopharmacol 36, 2169–2186 (2011). https://doi.org/10.1038/npp.2011.107
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/npp.2011.107
Keywords
This article is cited by
-
The CBP KIX domain regulates long-term memory and circadian activity
BMC Biology (2020)
-
Intracranial Self-Stimulation Modulates Levels of SIRT1 Protein and Neural Plasticity-Related microRNAs
Molecular Neurobiology (2020)
-
Jujuboside A prevents sleep loss-induced disturbance of hippocampal neuronal excitability and memory impairment in young APP/PS1 mice
Scientific Reports (2019)
-
AD-Related N-Terminal Truncated Tau Is Sufficient to Recapitulate In Vivo the Early Perturbations of Human Neuropathology: Implications for Immunotherapy
Molecular Neurobiology (2018)
-
Restoring synaptic plasticity and memory in mouse models of Alzheimer’s disease by PKR inhibition
Molecular Brain (2017)