Abstract
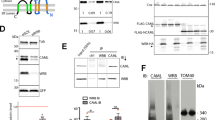
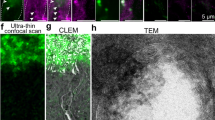
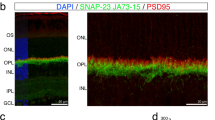
UNC119 is widely expressed among vertebrates and other phyla. We found that UNC119 recognized the acylated N terminus of the rod photoreceptor transducin α (Tα) subunit and Caenorhabditis elegans G proteins ODR-3 and GPA-13. The crystal structure of human UNC119 at 1.95-Å resolution revealed an immunoglobulin-like β-sandwich fold. Pulldowns and isothermal titration calorimetry revealed a tight interaction between UNC119 and acylated Gα peptides. The structure of co-crystals of UNC119 with an acylated Tα N-terminal peptide at 2.0 Å revealed that the lipid chain is buried deeply into UNC119′s hydrophobic cavity. UNC119 bound Tα-GTP, inhibiting its GTPase activity, thereby providing a stable UNC119–Tα-GTP complex capable of diffusing from the inner segment back to the outer segment after light-induced translocation. UNC119 deletion in both mouse and C. elegans led to G protein mislocalization. Thus, UNC119 is a Gα subunit cofactor essential for G protein trafficking in sensory cilia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Rosenbaum, J.L. & Witman, G.B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3, 813–825 (2002).
Jin, H. et al. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141, 1208–1219 (2010).
Kim, J., Krishnaswami, S.R. & Gleeson, J.G. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum. Mol. Genet. 17, 3796–3805 (2008).
Gillingham, A.K. & Munro, S. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 23, 579–611 (2007).
Zhang, H. et al. Deletion of PrBPδ impedes transport of GRK1 and PDE6 catalytic subunits to photoreceptor outer segments. Proc. Natl. Acad. Sci. USA 104, 8857–8862 (2007).
Keller, L.C. et al. Molecular architecture of the centriole proteome: the conserved WD40 domain protein POC1 is required for centriole duplication and length control. Mol. Biol. Cell 20, 1150–1166 (2009).
Chung, S., Kang, S., Paik, S. & Lee, J. NgUNC-119, Naegleria homologue of UNC-119, localizes to the flagellar rootlet. Gene 389, 45–51 (2007).
Maduro, M. & Pilgrim, D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141, 977–988 (1995).
Liu, Q. et al. The proteome of the mouse photoreceptor sensory cilium complex. Mol. Cell. Proteomics 6, 1299–1317 (2007).
Higashide, T., Murakami, A., McLaren, M.J. & Inana, G. Cloning of the cDNA for a novel photoreceptor protein. J. Biol. Chem. 271, 1797–1804 (1996).
Swanson, D.A., Chang, J.T., Campochiaro, P.A., Zack, D.J. & Valle, D. Mammalian orthologs of C. elegans unc-119 highly expressed in photoreceptors. Invest. Ophthalmol. Vis. Sci. 39, 2085–2094 (1998).
Ishiba, Y. et al. Targeted inactivation of synaptic HRG4 (UNC119) causes dysfunction in the distal photoreceptor and slow retinal degeneration, revealing a new function. Exp. Eye Res. 84, 473–485 (2007).
Higashide, T., McLaren, M.J. & Inana, G. Localization of HRG4, a photoreceptor protein homologous to Unc-119, in ribbon synapse. Invest. Ophthalmol. Vis. Sci. 39, 690–698 (1998).
Kobayashi, A., Kubota, S., Mori, N., McLaren, M.J. & Inana, G. Photoreceptor synaptic protein HRG4 (UNC119) interacts with ARL2 via a putative conserved domain. FEBS Lett. 534, 26–32 (2003).
Veltel, S., Kravchenko, A., Ismail, S. & Wittinghofer, A. Specificity of Arl2/Arl3 signaling is mediated by a ternary Arl3-effector-GAP complex. FEBS Lett. 582, 2501–2507 (2008).
Haeseleer, F. Interaction and colocalization of CaBP4 and Unc119 (MRG4) in photoreceptors. Invest. Ophthalmol. Vis. Sci. 49, 2366–2375 (2008).
Alpadi, K. et al. RIBEYE recruits Munc119, a mammalian ortholog of the Caenorhabditis elegans protein unc119, to synaptic ribbons of photoreceptor synapses. J. Biol. Chem. 283, 26461–26467 (2008).
Kobayashi, A. et al. HRG4 (UNC119) mutation found in cone-rod dystrophy causes retinal degeneration in a transgenic model. Invest. Ophthalmol. Vis. Sci. 41, 3268–3277 (2000).
Vepachedu, R., Karim, Z., Patel, O., Goplen, N. & Alam, R. Unc119 protects from Shigella infection by inhibiting the Abl family kinases. PLoS ONE 4, e5211 (2009).
Gorska, M.M., Cen, O., Liang, Q., Stafford, S.J. & Alam, R. Differential regulation of interleukin 5–stimulated signaling pathways by dynamin. J. Biol. Chem. 281, 14429–14439 (2006).
Bork, P., Holm, L. & Sander, C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 242, 309–320 (1994).
Hoffman, G.R., Nassar, N. & Cerione, R.A. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell 100, 345–356 (2000).
Hanzal-Bayer, M., Renault, L., Roversi, P., Wittinghofer, A. & Hillig, R.C. The complex of Arl2-GTP and PDE delta: from structure to function. EMBO J. 21, 2095–2106 (2002).
Zhang, H. et al. Photoreceptor cGMP phosphodiesterase delta subunit (PDEdelta) functions as a prenyl-binding protein. J. Biol. Chem. 279, 407–413 (2004).
Goc, A. et al. Different properties of the native and reconstituted heterotrimeric G protein transducin. Biochemistry 47, 12409–12419 (2008).
Kerov, V. et al. N-terminal fatty acylation of transducin profoundly influences its localization and the kinetics of photoresponse in rods. J. Neurosci. 27, 10270–10277 (2007).
Baehr, W., Morita, E., Swanson, R. & Applebury, M.L. Characterization of bovine rod outer segment G protein. J. Biol. Chem. 257, 6452–6460 (1982).
Marrari, Y., Crouthamel, M., Irannejad, R. & Wedegaertner, P.B. Assembly and trafficking of heterotrimeric G proteins. Biochemistry 46, 7665–7677 (2007).
Slepak, V.Z. & Hurley, J.B. Mechanism of light-induced translocation of arrestin and transducin in photoreceptors: interaction-restricted diffusion. IUBMB Life 60, 2–9 (2008).
Sokolov, M. et al. Massive light-driven translocation of transducin between the two major compartments of rod cells: a novel mechanism of light adaptation. Neuron 34, 95–106 (2002).
Elias, R.V., Sezate, S.S., Cao, W. & McGinnis, J.F. Temporal kinetics of the light/dark translocation and compartmentation of arrestin and alpha-transducin in mouse photoreceptor cells. Mol. Vis. 10, 672–681 (2004).
Calvert, P.D., Strissel, K.J., Schiesser, W.E., Pugh, E.N. Jr. & Arshavsky, V.Y. Light-driven translocation of signaling proteins in vertebrate photoreceptors. Trends Cell Biol. 16, 560–568 (2006).
Knobel, K.M., Davis, W.S., Jorgensen, E.M. & Bastiani, M.J. UNC-119 suppresses axon branching in C. elegans. Development 128, 4079–4092 (2001).
Roayaie, K., Crump, J.G., Sagasti, A. & Bargmann, C.I. The G alpha protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron 20, 55–67 (1998).
Lans, H., Rademakers, S. & Jansen, G. A network of stimulatory and inhibitory Galpha-subunits regulates olfaction in Caenorhabditis elegans. Genetics 167, 1677–1687 (2004).
Perkins, L.A., Hedgecock, E.M., Thomson, J.N. & Culotti, J.G. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117, 456–487 (1986).
Sokolov, M. et al. Phosducin facilitates light-driven transducin translocation in rod photoreceptors. Evidence from the phosducin knockout mouse. J. Biol. Chem. 279, 19149–19156 (2004).
Cowan, C.W., Wensel, T.G. & Arshavsky, V.Y. Enzymology of GTPase acceleration in phototransduction. Methods Enzymol. 315, 524–538 (2000).
Troemel, E.R., Kimmel, B.E. & Bargmann, C.I. Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell 91, 161–169 (1997).
Acton, T.B. et al. Robotic cloning and Protein Production Platform of the Northeast Structural Genomics Consortium. Methods Enzymol. 394, 210–243 (2005).
Chayen, N.E., Stewart, P.D., Maeder, D.L. & Blow, D.M. An automated system for micro-batch protein crystallization and screening. J. Appl. Crystallogr. 23, 297–302 (1990).
Otwinowski, Z. & Minor, D. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Schneider, T.R. & Sheldrick, G.M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 (2002).
Terwilliger, T.C. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374, 22–37 (2003).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–383 (2007).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Acknowledgements
We thank R. Abramowitz and J. Schwanof for access to the X4A beamline at the National Synchrotron Light Source and G. DeTitta of Hauptman Woodward Research Institute for crystallization screening. This work was supported by US National Institutes of Health grants EY08123 (W.B.), EY019298 (W.B.), EY014800-039003 (National Eye Institute core grant), EY10848 (G.I.) and NS034307 (E.M.J.), by the Howard Hughes Medical Institute (E.M.J.), by a grant from the Protein Structure Initiative of the US National Institutes of Health (U54 GM074958), by the University of Utah Macromolecule Crystallography Core Facility, by a Center Grant of the Foundation Fighting Blindness to the University of Utah, and unrestricted grants to the Departments of Ophthalmology at the University of Utah from Research to Prevent Blindness. W.B. is a recipient of a Research to Prevent Blindness Senior Investigator Award.
Author information
Authors and Affiliations
Contributions
H.Z. generated pulldown/light-induced translocation results and is responsible for C. elegans immunostaining and imaging. R.C. generated ITC results. R.C., F.G.W. and C.P.H. generated human UNC119/acylated Tα-peptide co-crystals and determined the structure. S.V., Y.C., J.S., Y.J.H., R.X., G.T.M. and L.T. determined the unbound human UNC119 structure. R.C., C.D.G. and W.B. isolated ROS membranes, transducin and determined GTPase activity. M.W.D. and E.M.J. generated transgenic C. elegans mutants. G.I. generated the Unc119 knockout mouse. H.Z., R.C., C.P.H., L.T. and W.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1 and 2 (PDF 2571 kb)
Rights and permissions
About this article
Cite this article
Zhang, H., Constantine, R., Vorobiev, S. et al. UNC119 is required for G protein trafficking in sensory neurons. Nat Neurosci 14, 874–880 (2011). https://doi.org/10.1038/nn.2835
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.2835
This article is cited by
-
Transport and barrier mechanisms that regulate ciliary compartmentalization and ciliopathies
Nature Reviews Nephrology (2024)
-
Mettl14-mediated m6A modification is essential for visual function and retinal photoreceptor survival
BMC Biology (2022)
-
Functional compartmentalization of photoreceptor neurons
Pflügers Archiv - European Journal of Physiology (2021)
-
Establishing and regulating the composition of cilia for signal transduction
Nature Reviews Molecular Cell Biology (2019)
-
Molecular determinants of Guanylate Cyclase Activating Protein subcellular distribution in photoreceptor cells of the retina
Scientific Reports (2018)