Abstract
QuantSeq provides an easy protocol to generate highly strand-specific next-generation sequencing (NGS) libraries close to the 3′ end of polyadenylated RNAs within 4.5 h. Only one fragment per transcript is generated, directly linking the number of reads mapping to a gene to its expression. QuantSeq reduces data analysis time and enables a higher level of multiplexing per run. QuantSeq is the RNA sample preparation method for accurate and affordable gene expression measurement.
Main
With the rapid development of NGS technologies, RNA-seq has become the new standard for transcriptome analysis. Although the price per base has been substantially reduced, sample preparation, sequencing and data processing are major cost factors in high-throughput screenings. QuantSeq reduces the expenditures in these areas.
Sample preparation. QuantSeq is a fast and easy protocol that generates NGS libraries of sequences close to the 3′ end of polyadenylated RNAs within 4.5 h with just 2 h of hands-on time. The kit requires only 0.5–500 ng of total RNA input without the need for poly(A) enrichment or ribosomal RNA depletion. Because of its focus on the 3′ end, QuantSeq is also highly suitable for formalin-fixed, paraffin-embedded samples.
Sequencing. QuantSeq generates only one fragment per transcript, and the number of reads mapped to a given gene is proportional to its expression. No complicated coverage-based quantification is required. Fewer reads are necessary for determining unambiguous gene-expression values, allowing a higher level of multiplexing.
Data processing. Most sequences will originate from the last exon and the 3′ untranslated region containing only a few splice junctions, dramatically reducing mapping time (6 samples in 35 min; for details see experiment below). QuantSeq's high strand specificity (>99.9%) enables the discovery and quantification of antisense transcripts and overlapping genes.
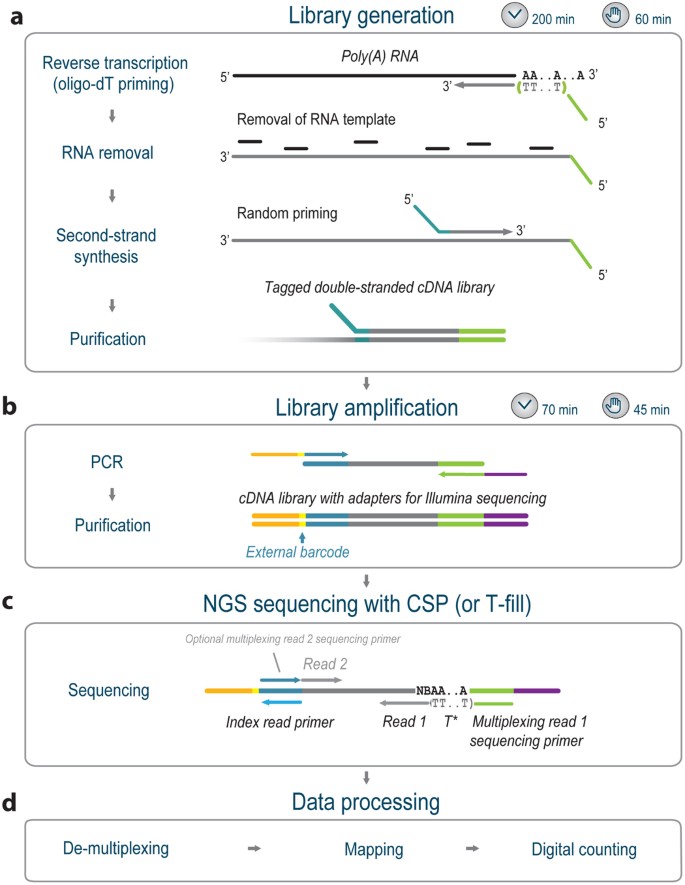
The QuantSeq workflow
Library generation is initiated by oligo-dT priming (Fig. 1a), and no prior poly(A) enrichment or ribosomal RNA depletion is required. First-strand synthesis and RNA removal is followed by random-primed synthesis of the complementary strand (second-strand synthesis). Illumina- or IonTorrent-specific linker sequences are introduced by the primers. The resulting double-stranded cDNA is purified with magnetic beads, rendering the protocol compatible with automation. Library PCR amplification then introduces the complete sequences required for cluster generation (Fig. 1b). Illumina libraries can be multiplexed with up to 96 external barcodes and are compatible with both single-end and paired-end sequencing reagents. The insert size is optimized for short reads (e.g., SR50 or SR100) while maintaining suitability for longer read lengths. IonTorrent libraries can be multiplexed using 24 in-line barcodes.
QuantSeq is available in two editions with different read orientations. The first edition, QuantSeq (cat. no. 015.24, 015.96 for Illumina and cat. no. 012.24 for IonTorrent), generates reads toward the poly(A) tail that correspond to the mRNA sequence during read 1 sequencing. Longer reads may be required if the exact 3′ end of the mRNA is of particular interest. The second edition, QuantSeq (T-fill) (cat. no. 016.24, 016.96 for Illumina only), generates reads corresponding to the cDNA sequence (Fig. 1c). Here, a customized sequencing primer (CSP) is used that covers the oligo(dT) stretch to achieve cluster calling on Illumina sequencers, which require a random base distribution within the first sequenced bases. Alternatively, a T-fill reaction can be carried out1.
Comparison between QuantSeq and standard mRNA sequencing
QuantSeq enables upscaling in multiplexing RNA-seq experiments, rendering it highly suitable for differential gene expression analysis. Here we present a comparison between QuantSeq and a standard mRNA-seq protocol, focusing on differential gene expression metrics.
We performed QuantSeq (T-fill) library preparations (cat. no. 016.24) on U.S. Food and Drug Administration (FDA) Sequencing Quality Control (SEQC) standard samples A and B in technical triplicates. Sample A is a mixture of Universal Human Reference RNA (UHRR) and External RNA Controls Consortium (ERCC) spike-in control mix 1. Sample B is a mixture of Human Brain Reference RNA (HBRR) and ERCC spike-in control mix 2 (we received SEQC samples A and B from the FDA prepared according to the FDA/National Center for Toxicological Research SEQC RNA Sample Preparation and Testing SOP_20110804). After T-fill, these 6 libraries, referred to as QuantSeq A1–3 and B1–3, were sequenced in one Illumina HiSeq 2000 lane yielding 150 M single reads of 50 bp (SR50). Residual adapter sequences were removed, and the trimmed pass-filter reads were down-sampled to 10 M each to be comparable with an mRNA-seq NGS experiment derived from the identical RNA input material. The mRNA-seq data sets were made available by a laboratory that participated in the recently published Association of Biomolecular Resource Facilities (ABRF) NGS study2. In that study, the researchers performed a stranded RNA-seq library preparation with poly(A) enrichment in 2 technical triplicates, obtaining 50 bp paired-end reads on an Illumina HiSeq 2000 (ref. 2; from the GSE48035 data set samples SRR903178–80 from GSM1166109 and SRR903210–12 from GSM1166113 were used in this comparison). We discarded read 2 in our 6 data sets, referred to as mRNA-seq A1–3 and B1–3, to obtain single-read data comparable to the QuantSeq data.
We pooled the 6 mRNA-seq data sets and aligned them to the GRCh 37.73 genome assembly including ERCC sequences using a splice-junction mapper, TopHat2, which required 2 h 50 min. Notably, the pooled 6 QuantSeq data sets were aligned in only 35 min using the short read aligner Bowtie2 on the same computer system. For gene expression quantification, standard mRNA-seq relies on length normalization of the number of reads to fragments per kilobase of exon per million fragments mapped, which depends on the correctness of read-to-transcript assignments carried out by Cufflinks. As QuantSeq generates only one fragment per transcript, length normalization is not required, and gene expression quantification is read-count based (Fig. 1d). Mapped reads were further categorized with htseq-count (Table 1).
Data sets were evaluated for ERCC spike-in abundances. QuantSeq detected the actual amount of ERCC RNAs that was spiked in (3% relative to 2% mRNA in total RNA). In the same input RNA, the standard mRNA-seq experiment detected only 1% ERCC sequences. This underrepresentation is most likely caused by a less efficient poly(A) selection of the spike-in RNA's short poly(A) tails. To allow a direct comparison, all ERCC reads were down-sampled to identical ERCC read numbers. These subsets of ERCC reads were processed with routines embedded in the recently released ERCC dashboard3.
One major benefit of QuantSeq can be visualized by plotting the relative coverage across the normalized transcript length (Fig. 2). Standard mRNA-seq distributes reads across the entire length of transcripts with underrepresentation of 3′ and 5′ ends, whereas QuantSeq covers the very 3′ end of transcripts. In fact, for gene expression and differential expression analysis, one read per transcript is sufficient. The additional sequencing space gained by focusing on the 3′ end can be used for a higher degree of multiplexing. In the present example, standard mRNA-seq has a 12.4-fold higher relative sequence coverage (area under the curve (AUC) ratio for all genes (Fig. 2)), which in turn presents the maximal possible reduction in read depth when using QuantSeq while still determining gene expression accurately.
We compared the results from the QuantSeq and mRNA-seq experiments focusing on differential gene expression3. The ability of a method to measure differentials can be evaluated using the predetermined fold changes between ERCC spike-in control mixes 1 and 2. When plotting the true-positive rate versus the false-positive rate, the AUC is a measure for the correct detection of differential gene expression (Fig. 3). The maximum mean AUC value, corresponding to optimal differential detection, is 1. When the number of reads is down-sampled from 10 M to 0.625 M, standard mRNA-seq obtains mean AUC values of around 0.72 only, whereas QuantSeq maintains very high AUC values of around 0.90, although similar total numbers of ERCC spike-in RNAs were detected by both methods during the course of down-sampling.
The predetermined fold changes between ERCC spike-in control mixes 1 and 2 were used to assess TPRs and FPRs. The receiver operator characteristic–derived AUC value is one measure for the correct detection of differential gene expression. AUC values were assessed together with the number of ERCC RNAs detected (#ERCC) for reads down-sampled from 10 M to 0.625 M. The averaged values of the 6 samples A1–3 and B1–3 each are presented in the insert table. PF, past filter.
Conclusions
QuantSeq is a robust and simple mRNA sequencing method. It increases the precision in gene expression measurements as only one read per transcript is generated. At lower read depths, such focus on the 3′ end results in higher stability of differential gene expression measurements. QuantSeq is ideal for increasing the degree of multiplexing in NGS gene expression experiments and is the method of choice for accurately determining gene expression at the lowest cost.
Addendum
QuantSeq is one kit of a series of transcriptome analysis kits provided by Lexogen. For a highly efficient extraction of either total RNA or split fractions of large and small RNA, we offer the SPLIT RNA Extraction Kit (cat. no. 008.48). Complementary to the 3′ end–focused QuantSeq kit, the SENSE Total RNA library preparation kit (cat. no. 009.08-96) and the SENSE mRNA-seq library preparation kit (versions available for Illumina, Ion Torrent and Solid) provide transcript body–covering RNA-seq libraries of superior strand specificity in less than 5 h. For alternative applications such as promoter and polyadenylation analysis, splice variant determination, probe generation, etc., the TeloPrime Full-Length cDNA Amplification Kit (cat. no. 013.04-24) generates full-length cDNA libraries with precisely tagged start and end sites.
References
Wilkening, S. et al. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res. 41, e65 (2013).
Li, S. et al. Multi-platform assessment of transcriptome profiling using RNA-seq in the ABRF next-generation sequencing study. Nat. Biotechnol. 32, 915–925 (2014).
Munro, S.A. et al. Assessing technical performance in differential gene expression experiments with external spike-in RNA control ratio mixtures. Nat. Commun. 5, 5125 (2014).
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclaimer
This article was submitted to Nature Methods by a commercial organization and has not been peer reviewed. Nature Methods takes no responsibility for the accuracy or otherwise of the information provided.
Rights and permissions
About this article
Cite this article
Moll, P., Ante, M., Seitz, A. et al. QuantSeq 3′ mRNA sequencing for RNA quantification. Nat Methods 11, i–iii (2014). https://doi.org/10.1038/nmeth.f.376
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.f.376
This article is cited by
-
The forgotten variable? Does the euthanasia method and sample storage condition influence an organisms transcriptome – a gene expression analysis on multiple tissues in pigs
BMC Genomics (2023)
-
The NELF pausing checkpoint mediates the functional divergence of Cdk9
Nature Communications (2023)
-
Transcriptomic profiling implicates PAF1 in both active and repressive immune regulatory networks
BMC Genomics (2022)
-
Simultaneous metabolite MALDI-MSI, whole exome and transcriptome analysis from formalin-fixed paraffin-embedded tissue sections
Laboratory Investigation (2022)
-
Transcriptomic networks of gba3 governing specification of the dopaminergic neurons in zebrafish embryos
Genes & Genomics (2022)