Abstract
Microglia are damage sensors for the central nervous system (CNS), and the phagocytes responsible for routine non-inflammatory clearance of dead brain cells1. Here we show that the TAM receptor tyrosine kinases Mer and Axl2 regulate these microglial functions. We find that adult mice deficient in microglial Mer and Axl exhibit a marked accumulation of apoptotic cells specifically in neurogenic regions of the CNS, and that microglial phagocytosis of the apoptotic cells generated during adult neurogenesis3,4 is normally driven by both TAM receptor ligands Gas6 and protein S5. Using live two-photon imaging, we demonstrate that the microglial response to brain damage is also TAM-regulated, as TAM-deficient microglia display reduced process motility and delayed convergence to sites of injury. Finally, we show that microglial expression of Axl is prominently upregulated in the inflammatory environment that develops in a mouse model of Parkinson’s disease6. Together, these results establish TAM receptors as both controllers of microglial physiology and potential targets for therapeutic intervention in CNS disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ransohoff, R. M. & Cardona, A. E. The myeloid cells of the central nervous system parenchyma. Nature 468, 253–262 (2010)
Lemke, G. Biology of the TAM receptors. Cold Spring Harb. Perspect. Biol. 5, a009076 (2013)
Aimone, J. B. et al. Regulation and function of adult neurogenesis: from genes to cognition. Physiol. Rev. 94, 991–1026 (2014)
Sierra, A. et al. Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell 7, 483–495 (2010)
Lew, E. D. et al. Differential TAM receptor-ligand-phospholipid interactions delimit differential TAM bioactivities. eLife 3, e03385 (2014)
Chandra, S., Gallardo, G., Fernandez-Chacon, R., Schluter, O. M. & Sudhof, T. C. α-Synuclein cooperates with CSPα in preventing neurodegeneration. Cell 123, 383–396 (2005)
Ginhoux, F. et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010)
Lu, Q. & Lemke, G. Homeostatic regulation of the immune system by receptor tyrosine kinases of the Tyro 3 family. Science 293, 306–311 (2001)
Rothlin, C. V., Ghosh, S., Zuniga, E. I., Oldstone, M. B. & Lemke, G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell 131, 1124–1136 (2007)
Burstyn-Cohen, T. et al. Genetic dissection of TAM receptor-ligand interaction in retinal pigment epithelial cell phagocytosis. Neuron 76, 1123–1132 (2012)
Scott, R. S. et al. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature 411, 207–211 (2001)
Zagórska, A., Través, P. G., Lew, E. D., Dransfield, I. & Lemke, G. Diversification of TAM receptor tyrosine kinase function. Nature Immunol. 15, 920–928 (2014)
Bhattacharyya, S. et al. Enveloped viruses disable innate immune responses in dendritic cells by direct activation of TAM receptors. Cell Host Microbe 14, 136–147 (2013)
Zhang, Z. et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nature Genet. 44, 852–860 (2012)
Lai, C. & Lemke, G. An extended family of protein-tyrosine kinase genes differentially expressed in the vertebrate nervous system. Neuron 6, 691–704 (1991)
Prieto, A. L., O’Dell, S., Varnum, B. & Lai, C. Localization and signaling of the receptor protein tyrosine kinase Tyro3 in cortical and hippocampal neurons. Neuroscience 150, 319–334 (2007)
Gautier, E. L. et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature Immunol. 13, 1118–1128 (2012)
Grommes, C. et al. Regulation of microglial phagocytosis and inflammatory gene expression by Gas6 acting on the Axl/Mer family of tyrosine kinases. J. Neuroimmune Pharmacol. 3, 130–140 (2008)
Ji, R. et al. TAM receptors affect adult brain neurogenesis by negative regulation of microglial cell activation. J. Immunol. 191, 6165–6177 (2013)
Cardona, A. E. et al. Control of microglial neurotoxicity by the fractalkine receptor. Nature Neurosci. 9, 917–924 (2006)
Ito, D. et al. Microglia-specific localisation of a novel calcium binding protein, Iba1. Brain Res. Mol. Brain Res. 57, 1–9 (1998)
Jung, S. et al. Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20, 4106–4114 (2000)
Chung, W. S. et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature 504, 394–400 (2013)
Parkhurst, C. N. et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155, 1596–1609 (2013)
Miner, J. J. et al. The TAM receptor Mertk protects against neuroinvasive viral infection by maintaining blood-brain barrier integrity. Nature Med. 21, 1464–1472 (2015)
Brown, G. C. & Neher, J. J. Microglial phagocytosis of live neurons. Nature Rev. Neurosci. 15, 209–216 (2014)
Burstyn-Cohen, T., Heeb, M. J. & Lemke, G. Lack of protein S in mice causes embryonic lethal coagulopathy and vascular dysgenesis. J. Clin. Invest. 119, 2942–2953 (2009)
Nimmerjahn, A., Kirchhoff, F. & Helmchen, F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318 (2005)
Tang, Y. et al. Mertk deficiency affects macrophage directional migration via disruption of cytoskeletal organization. PLoS ONE 10, e0117787 (2015)
Mattsson, N. et al. CSF protein biomarkers predicting longitudinal reduction of CSF β-amyloid42 in cognitively healthy elders. Transl. Psychiatry 3, e293 (2013)
Lu, Q. et al. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature 398, 723–728 (1999)
Angelillo-Scherrer, A. et al. Deficiency or inhibition of Gas6 causes platelet dysfunction and protects mice against thrombosis. Nature Med. 7, 215–221 (2001)
Vives, V., Alonso, G., Solal, A. C., Joubert, D. & Legraverend, C. Visualization of S100B-positive neurons and glia in the central nervous system of EGFP transgenic mice. J. Comp. Neurol. 457, 404–419 (2003)
Chung, K. et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013)
Miksa, M., Komura, H., Wu, R., Shah, K. G. & Wang, P. A novel method to determine the engulfment of apoptotic cells by macrophages using pHrodo succinimidyl ester. J. Immunol. Methods 342, 71–77 (2009)
Dransfield, I., Zagorska, A., Lew, E. D., Michail, K. & Lemke, G. Mer receptor tyrosine kinase mediates both tethering and phagocytosis of apoptotic cells. Cell Death Dis. 6, e1646 (2015)
Sharma, K. et al. Cell type- and brain region-resolved mouse brain proteome. Nature Neurosci. 18, 1819–1831 (2015)
Drew, P. J. et al. Chronic optical access through a polished and reinforced thinned skull. Nature Methods 7, 981–984 (2010)
Knowland, D. et al. Stepwise recruitment of transcellular and paracellular pathways underlies blood-brain barrier breakdown in stroke. Neuron 82, 603–617 (2014)
Acknowledgements
This work was supported by grants from the US National Institutes of Health (R01 NS085296 and R01 AI101400 to G.L., DP2 NS083038 and R01 NS085938 to A.N., R01 AI089824 to C.V.R., and P30CA014195 to the Salk Institute), the Leona M. and Harry B. Helmsley Charitable Trust (#2012-PG-MED002 to the Salk Institute), the Nomis, H. N. and Frances C. Berger, Fritz B. Burns, and HKT Foundations (to G.L.), and the Waitt, Rita Allen and Hearst Foundations (to A.N.); and by postdoctoral fellowships from the Marie Curie International Outgoing Fellowship Program (to P.G.T.), the Nomis Foundation (to A.Z. and E.D.L.), and the Howard Hughes Medical Institute Life Sciences Research Foundation (to Y.T.). We thank J. Hash for excellent technical assistance and J. Flynn for help with the CLARITY method.
Author information
Authors and Affiliations
Contributions
L.F. and P.G.T. designed experiments, performed apoptotic cell, BrdU, immunohistochemical, and genetic analyses, and contributed equally to the paper; Y.T., L.F. and H.L.-B. performed and analysed in vivo two photon imaging; E.D.L. prepared TAM ligands; P.G.B. performed brain histology; P.C. analysed cytokine profiles; A.Z. analysed Axl expression in Parkinson’s disease transgenics; C.V.R. provided floxed Mertk alleles; A.N. designed and implemented two-photon imaging; and G.L. designed experiments and wrote the paper. All authors edited the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Mer is expressed by microglia.
a, Brain (hippocampus) sections from Cx3cr1GFP/+ mice that were wild-type (top row) or Axl−/−Mertk−/− (bottom row) were visualized by confocal microscopy for GFP (1st column), anti-Mer (red, 2nd column), or anti-GFAP (cyan, 3rd column) immunoreactivity; 4th column, merged images. Scale bars, 10 μm. Axl immunostaining signal is too low to be visualized in unactivated microglia (not shown; but see Fig. 4d). b, Mer expression does not co-localize with S100b+ cells. Immunostaining of Cx3cr1GFP/+ brain sections with anti-Mer (red, 2nd panel) and anti-S100b (cyan, 3rd panel); 4th panel, merged images. c, Mer co-localizes with Iba1, but not GFAP or GFP in S100bGFP/+ mice. Brain sections were visualized by confocal microscopy for anti-Iba1 (top) or anti-Gfap (bottom) (both cyan, 1st column), anti-Mer (red, 2nd column), or GFP (green, 3rd column) immunoreactivity; 4th column, merged images. Scale bars (b, c), 20 μm. Representative images from analyses performed in n = 2 mice (a–c).
Extended Data Figure 2 Accumulation of apoptotic cells is confined to neurogenic and derivative migratory regions of the Axl−/−Mertk−/− CNS.
a, A low power tiled image of a section through the Axl−/−Mertk−/− subventricular zone and surrounding brain tissue, stained for cCasp3, illustrates that apoptotic cells are confined within the SVZ. b, A low power tiled image of a section through the Axl−/−Mertk−/− rostral migratory stream (RMS) and surrounding brain tissue illustrates that cCasp3+ apoptotic cells are confined within the RMS. c, A low power tiled image of the granule cell and mitral cell layers (gcl and mcl, respectively) of the Axl−/−Mertk−/− olfactory bulb, stained for cCasp3, illustrates that there are no apoptotic cells detected in the double mutant bulb. Scale bars (a–c), 200 μm. Representative images from analyses performed in n = 3 mice (a–c).
Extended Data Figure 3 Mer is the principal microglial TAM receptor required for phagocytosis of apoptotic cells in the SVZ.
a, Sections of the SVZ from Axl−/− (top row) and Mertk−/− (bottom row) mice immunostained for cCasp3 and NeuN (green and red, respectively; left panels), or Iba1 and NeuN (green and red, respectively; right panels) reveal the accumulation of cCasp3+ apoptotic cells only in the Mertk−/− SVZ. b, Sections of the SVZ (top) and RMS (bottom) of Gas6−/− mice, illustrating no accumulation of apoptotic cells (similar to both wild type and Axl−/−). c, Sections of the SVZ (top) and RMS (bottom) of Mertk−/−Gas6−/− mice, illustrating a massive accumulation of aoptotic cells, similar to that seen in Axl−/−Mertk−/− mice. Scale bars, 50 μm. See main text for quantification. Representative images from analyses performed in n = 2 mice for Gas6−/− and Mertk−/−Gas6−/−, and n = 3 mice for Axl−/− and Mertk−/−.
Extended Data Figure 4 Conditional Mertk knockouts.
The knockout strategy targets exon 18 of the wild-type mouse Mertk gene, which encodes residues W779–L824 of the tyrosine kinase domain (1st line). Deletion of this exon leads to a functional and null protein (see Methods, and Extended Data Fig. 5). The targeting vector (2nd line) had a PGK-Neo cassette for selection in embryonic stem (ES) cells, and contained loxP and FRT sites, recognized by Cre and Flp recombinases, respectively, at the indicated positions. Five ES cell lines with homologous recombination at the Mertk locus were identified by Southern blots of MfeI-digested DNA, using the indicated Pb 1/2 (external) and Pb 3/4 (internal) probes (3rd line). Introduction of Flp recombinase, achieved by crossing high percentage chimaeras (obtained from blastocyst injection of these ES cells) to C57Bl/6 FLP mice, removed the Neo cassette, leaving exon 18 flanked by loxP sites (4th line). Cre-mediated recombination at these loxP sites deletes exon 18. Mertkfl/fl mice, together with PCR-based protocols for their genotyping, are available upon request from the Rothlin laboratory (contact C.V.R.). See Methods for further information.
Extended Data Figure 5 Persistence of microglial-specific Mer ablation following tamoxifen injection of Cx3cr1CreER/+Mertkfl/fl mice.
a, Mice were injected intraperitoneally with oil vehicle alone (−tamoxifen, top row) or with tamoxifen (+tamoxifen, bottom row) (see Methods), and brain sections were immunostained for Mer protein expression (red panels in 2nd, 4th columns) in Iba1+ microglia (green panels in 1st, 3rd columns) at 1 week (left four panels) and 7 weeks (right four panels) after injection. Sections counter-stained with Hoechst 33258 to visualize nuclei (blue). b, Brain sections containing a brain capillary 7 weeks after injection of vehicle (top) or tamoxifen (bottom), showing that although Mer expression in microglia is eliminated upon tamoxifen-mediated Cx3cr1-restricted induction of Cre activity, Mer expression in CD31+ microvascular endothelial cells (arrows) is maintained. Representative images of n = 2 mice per time point.
Extended Data Figure 6 Identity of immigrant BrdU+ cells in the olfactory bulb.
a, A group of the BrdU+ cells in the glomerular layer (gl), visualized 35 days after injection of BrdU (red) and presumed immigrant descendants of SVZ cells in S phase at the time of injection, are also positive for tyrosine hydroxylase (TH, green) in both wild-type (left panel) and Axl−/−Mertk−/− (right panel) mice. Arrowheads are examples of TH+BrdU+ cells. b, Similar comparative granule cell layer (gcl) sections stained with anti-BrdU (red) and calretinin (CR, green). Arrowheads are examples of CR+BrdU+ cells. Sections were co-stained with Hoechst 33258 to visualize nuclei. Scale bars, 50 μm. Representative images of n = 2 per genotype.
Extended Data Figure 7 Both Gas6 and Pros1 drive microglial phagocytosis of apoptotic cells in vitro.
a, Cultured microglia express Mer but little or no Axl under basal conditions. Microglia were cultured from wild-type Cx3cr1GFP/+ mice, visualized for GFP (1st column), and immunostained for Iba1 (3rd column), Mer (2nd column, top), and Axl (2nd column, bottom). Scale bar, 10 μm. b–d, In vitro pHrodo-based assay of phagocytosis of apoptotic cells by microglia (see Methods). b, In serum-containing medium (10% FBS), wild-type microglia are effective phagocytes; mean phagocytic activity is substantially reduced in Axl−/−Mertk−/− (A−/−M−/−) microglia. c, d, Both purified Gas6 (c) and purified Pros1 (d) stimulate AC phagocytosis by cultured microglia in serum-free medium, and this stimulation is entirely TAM-dependent. e, The phagocytic activity of cultured astroglia prepared from Cx3cr1GFP/+ mice that were either wild-type or Axl−/−Mertk−/− was measured in the same pHrodo-based assay in serum-free medium ± Gas6. For this FACS-based assay, astrocytes were gated using an astrocyte-specific surface antigen-2 (ACSA-2) antibody (see Methods). Bar graphs represent mean phagocytic activity (± s.em.); n = 2 replicates from 2 mice per genotype (b–d), and 2 replicates from 4 mice per genotype (e).
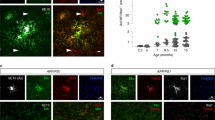
Extended Data Figure 8 Regulation of microglial Axl by neuroinflammation.
a, b, Axl (a) and Mer (b) regulation in purified (GFP+) cultured microglia by the tolerogenic stimulus dexamethasone (Dex) and the two proinflammatory stimuli IFNγ and poly(I:C), as assessed by immunostaining. Axl expression (a) is very low in the absence of an added stimulus, is not elevated by Dex, but is strongly upregulated by both IFNγ and poly(I:C). In contrast, Mer expression (b) is readily detected in the absence of an added stimulus, is further elevated by Dex, but is modestly suppressed by both IFNγ and poly(I:C). Scale bar, 10 μm. c, In contrast to the spinal cord (see Fig. 4a), there is no upregulation of the indicated inflammatory mediator/marker mRNAs (mean expression ± s.e.m.) in the spleens, and only modest upregulation in the brains, of SNCAA53T mice at 8–10 months of age. n = 3 mice for each genotype. d, Western blot analysis of spleen (left blots) and brain and spinal cord (right blots) extracts from two different wild-type mice and four or three different SNCAA53T mice at 9–10 months, for the indicated proteins, with Gapdh as a loading control. Note that soluble Axl ectodomain (sAxl) is upregulated in the SNCAA53T spinal cord concomitantly with Axl. e, Although Axl is strongly upregulated in Iba1+ microglia in the SNCAA53T spinal cord (see Fig. 4d), no upregulation of Mer is observed in these same cells. Scale bar, 10 μm. n = 2 wild-type and 3 SNCAA53T mice.
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1 - original source images for western blots for Figure 4 and Extended Data Figure 8 (the boxes delimit cropped areas used in the figures) and Supplementary Table 1, a list of quantitative PCR primers used in this paper. (PDF 1255 kb)
Microglia distribution in the SVZ and brain.
This sequence moves through a 1 mm3 block of fixed tissue of a Cx3cr1GFP/+ mouse brain, cleared using the CLARITY protocol (see Methods), and visualized for GFP (green), which is expressed by microglia. The sequence begins in the lateral ventricle (the black region at the center and left of the image) and extends into the SVZ as the video progresses. Note the regular tiling of the brain/SVZ parenchyma by evenly spaced GFP+ microglia. The video was generated by compiling successive confocal planes through the CLARITY-cleared tissue block. (MOV 5387 kb)
Process extension by wild-type microglia.
This sequence is a tracking series from a live head-restrained mouse, using two-photon imaging of GFP+ microglial processes (white) of Cx3cr1GFP/+ visual cortex, with the movement of 13 individual processes highlighted in colors over time (total video is ~67 minutes). (MOV 2557 kb)
Process extension by Axl-/-Mertk-/- microglia.
This sequence is a tracking series from a live head-restrained mouse, using a live two-photon imaging of GFP+ microglial processes (white) of Cx3cr1GFP/+Axl-/-Mertk-/- visual cortex, with the movement of 13 individual processes highlighted in colors over time (total video is ~63 minutes). (MOV 1769 kb)
Process extension toward lesion by wild-type microglia
This sequence is a tracking series from a live head-restrained mouse, using two-photon imaging of GFP+ microglial processes (white) of Cx3cr1GFP/+ visual cortex, with the movement of 10 individual processes toward a laser-induced microvascular lesion site (generated at time 0) highlighted in colors over time (total video ~11 minutes). (MOV 159 kb)
Process extension toward lesion by Axl-/-Mertk-/- microglia.
This sequence is a tracking series from a live head-restrained mouse, using two-photon imaging of GFP+ microglial processes (white) of Cx3cr1GFP/+Axl-/-Mertk-/- visual cortex, with the movement of 10 individual processes toward a laser-induced microvascular lesion site (generated at time 0) highlighted in colors over time (total video is ~12 minutes). (MOV 335 kb)
Rights and permissions
About this article
Cite this article
Fourgeaud, L., Través, P., Tufail, Y. et al. TAM receptors regulate multiple features of microglial physiology. Nature 532, 240–244 (2016). https://doi.org/10.1038/nature17630
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature17630
This article is cited by
-
Cell death by phagocytosis
Nature Reviews Immunology (2024)
-
Different inflammatory signatures based on CSF biomarkers relate to preserved or diminished brain structure and cognition
Molecular Psychiatry (2024)
-
Mertk-expressing microglia influence oligodendrogenesis and myelin modelling in the CNS
Journal of Neuroinflammation (2023)
-
Microglial function, INPP5D/SHIP1 signaling, and NLRP3 inflammasome activation: implications for Alzheimer’s disease
Molecular Neurodegeneration (2023)
-
Understanding microglial responses in large animal models of traumatic brain injury: an underutilized resource for preclinical and translational research
Journal of Neuroinflammation (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.