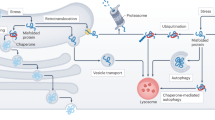
Abstract
Most proteins must fold into defined three-dimensional structures to gain functional activity. But in the cellular environment, newly synthesized proteins are at great risk of aberrant folding and aggregation, potentially forming toxic species. To avoid these dangers, cells invest in a complex network of molecular chaperones, which use ingenious mechanisms to prevent aggregation and promote efficient folding. Because protein molecules are highly dynamic, constant chaperone surveillance is required to ensure protein homeostasis (proteostasis). Recent advances suggest that an age-related decline in proteostasis capacity allows the manifestation of various protein-aggregation diseases, including Alzheimer's disease and Parkinson's disease. Interventions in these and numerous other pathological states may spring from a detailed understanding of the pathways underlying proteome maintenance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Dobson, C. M., Sali, A. & Karplus, M. Protein folding — a perspective from theory and experiment. Angew. Chem. Int. Edn Engl. 37, 868–893 (1998).
Bartlett, A. I. & Radford, S. E. An expanding arsenal of experimental methods yields an explosion of insights into protein folding mechanisms. Nature Struct. Mol. Biol. 16, 582–588 (2009).
Dunker, A. K., Silman, I., Uversky, V. N. & Sussman, J. L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 18, 756–764 (2008).
Powers, E. T., Morimoto, R. I., Dillin, A., Kelly, J. W. & Balch, W. E. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 78, 959–991 (2009).
Morimoto, R. I. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 22, 1427–1438 (2008).
Balch, W. E., Morimoto, R. I., Dillin, A. & Kelly, J. W. Adapting proteostasis for disease intervention. Science 319, 916–919 (2008).
Hartl, F. U. Molecular chaperones in cellular protein folding. Nature 381, 571–580 (1996).
Kubelka, J., Hofrichter, J. & Eaton, W. A. The protein folding 'speed limit'. Curr. Opin. Struct. Biol. 14, 76–88 (2004).
Herbst, R., Schafer, U. & Seckler, R. Equilibrium intermediates in the reversible unfolding of firefly (Photinus pyralis) luciferase. J. Biol. Chem. 272, 7099–7105 (1997).
Ellis, R. J. & Minton, A. P. Protein aggregation in crowded environments. Biol. Chem. 387, 485–497 (2006).
Tokuriki, N. & Tawfik, D. S. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature 459, 668–671 (2009).
Rutherford, S. L. & Lindquist, S. Hsp90 as a capacitor for morphological evolution. Nature 396, 336–342 (1998). This seminal study puts forward the idea that chaperones function in buffering the otherwise deleterious consequences of mutations.
Skach, W. R. Cellular mechanisms of membrane protein folding. Nature Struct. Mol. Biol. 16, 606–612 (2009).
Kerner, M. J. et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli . Cell 122, 209–220 (2005).
Eichner, T., Kalverda, A. P., Thompson, G. S., Homans, S. W. & Radford, S. E. Conformational conversion during amyloid formation at atomic resolution. Mol. Cell 41, 161–172 (2011). This exciting paper describes, at atomic resolution, the structural features of a non-native folding intermediate that are critical for amyloidogenic aggregation.
Chiti, F. & Dobson, C. M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006).
Bolognesi, B. et al. ANS binding reveals common features of cytotoxic amyloid species. ACS Chem. Biol. 5, 735–740 (2010) This paper provides evidence that the exposure of hydrophobic surfaces by oligomeric aggregation intermediates correlates with their toxicity.
Kayed, R. et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 (2003).
Hartl, F. U. & Hayer-Hartl, M. Converging concepts of protein folding in vitro and in vivo . Nature Struct. Mol. Biol. 16, 574–581 (2009).
Langer, T. et al. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356, 683–689 (1992).
Frydman, J., Nimmesgern, E., Ohtsuka, K. & Hartl, F. U. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature 370, 111–117 (1994).
Ellis, R. J. Molecular chaperones: assisting assembly in addition to folding. Trends Biochem. Sci. 31, 395–401 (2006).
Liu, C. et al. Coupled chaperone action in folding and assembly of hexadecameric Rubisco. Nature 463, 197–202 (2010).
Auluck, P. K., Chan, H. Y. E., Trojanowski, J. Q., Lee, V. M. Y. & Bonini, N. M. Chaperone suppression of α-synuclein toxicity in a Drosophila model for Parkinson's disease. Science 295, 865–868 (2002).
Mayer, M. P. Gymnastics of molecular chaperones. Mol. Cell 39, 321–331 (2010).
Kampinga, H. H. & Craig, E. A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nature Rev. Mol. Cell Biol. 11, 579–592 (2010).
Arndt, V. et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr. Biol. 20, 143–148 (2010).
Rüdiger, S., Buchberger, A. & Bukau, B. Interaction of Hsp70 chaperones with substrates. Nature Struct. Biol. 4, 342–349 (1997). A key paper in understanding how chaperones recognize their substrates.
Rousseau, F., Serrano, L. & Schymkowitz, J. W. H. How evolutionary pressure against protein aggregation shaped chaperone specificity. J. Mol. Biol. 355, 1037–1047 (2006).
Sharma, S. K., De los Rios, P., Christen, P., Lustig, A. & Goloubinoff, P. The kinetic parameters and energy cost of the Hsp70 chaperone as a polypeptide unfoldase. Nature Chem. Biol. 6, 914–920 (2010).
Frydman, J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70, 603–647 (2001).
Horwich, A. L. & Fenton, W. A. Chaperonin-mediated protein folding: using a central cavity to kinetically assist polypeptide chain folding. Q. Rev. Biophys. 42, 83–116 (2009).
Fujiwara, K., Ishihama, Y., Nakahigashi, K., Soga, T. & Taguchi, H. A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 29, 1552–1564 (2010).
Raineri, E., Ribeca, P., Serrano, L. & Maier, T. A more precise characterization of chaperonin substrates. Bioinformatics 26, 1685–1689 (2010).
Tartaglia, G. G., Dobson, C. M., Hartl, F. U. & Vendruscolo, M. Physicochemical determinants of chaperone requirements. J. Mol. Biol. 400, 579–588 (2010).
Xu, Z. H., Horwich, A. L. & Sigler, P. B. The crystal structure of the asymmetric GroEL−GroES−(ADP)7 chaperonin complex. Nature 388, 741–749 (1997).
Brinker, A. et al. Dual function of protein confinement in chaperonin-assisted protein folding. Cell 107, 223–233 (2001).
Tang, Y. C. et al. Structural features of the GroEL−GroES nano-cage required for rapid folding of encapsulated protein. Cell 125, 903–914 (2006).
Chakraborty, K. et al. Chaperonin-catalyzed rescue of kinetically trapped states in protein folding. Cell 142, 112–122 (2010).
Thirumalai, D. & Lorimer, G. H. Chaperonin-mediated protein folding. Annu. Rev. Biophys. Biomol. Struct. 30, 245–269 (2001).
Lin, Z., Madan, D. & Rye, H. S. GroEL stimulates protein folding through forced unfolding. Nature Struct. Mol. Biol. 15, 303–311 (2008).
Sharma, S. et al. Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell 133, 142–153 (2008).
Munoz, I. G. et al. Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nature Struct. Mol. Biol. 18, 14–19 (2011).
Douglas, N. R. et al. Dual action of ATP hydrolysis couples lid closure to substrate release into the Group II chaperonin chamber. Cell 144, 240–252 (2011).
Reissmann, S., Parnot, C., Booth, C. R., Chiu, W. & Frydman, J. Essential function of the built-in lid in the allosteric regulation of eukaryotic and archaeal chaperonins. Nature Struct. Mol. Biol. 14, 432–440 (2007).
Kitamura, A. et al. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nature Cell Biol. 8, 1163–1170 (2006).
Behrends, C. et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol. Cell 23, 887–897 (2006).
Tam, S., Geller, R., Spiess, C. & Frydman, J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nature Cell Biol. 8, 1155–1162 (2006).
Taipale, M., Jarosz, D. F. & Lindquist, S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nature Rev. Mol. Cell Biol. 11, 515–528 (2010).
McClellan, A. J. et al. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell 131, 121–135 (2007).
Scheufler, C. et al. Structure of TPR domain–peptide complexes: critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell 101, 199–210 (2000).
Wandinger, S. K., Richter, K. & Buchner, J. The Hsp90 chaperone machinery. J. Biol. Chem. 283, 18473–18477 (2008).
Ali, M. M. U. et al. Crystal structure of an Hsp90–nucleotide–p23/Sba1 closed chaperone complex. Nature 440, 1013–1017 (2006).
Shiau, A. K., Harris, S. F., Southworth, D. R. & Agard, D. A. Structural analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell 127, 329–340 (2006).
Neckers, L. Heat shock protein 90: the cancer chaperone. J. Biosci. 32, 517–530 (2007).
Geller, R., Vignuzzi, M., Andino, R. & Frydman, J. Evolutionary constraints on chaperone-mediated folding provide an antiviral approach refractory to development of drug resistance. Genes Dev. 21, 195–205 (2007). This seminal study describes the requirement of HSP90 in viral assembly, outlining a strategy for antiviral treatment based on HSP90 inhibition.
Eichmann, C., Preissler, S., Riek, R. & Deuerling, E. Cotranslational structure acquisition of nascent polypeptides monitored by NMR spectroscopy. Proc. Natl Acad. Sci. USA 107, 9111–9116 (2010).
Cabrita, L. D., Hsu, S. T., Launay, H., Dobson, C. M. & Christodoulou, J. Probing ribosome-nascent chain complexes produced in vivo by NMR spectroscopy. Proc. Natl Acad. Sci. USA 106, 22239–22244 (2009).
Lu, J. L. & Deutsch, C. Folding zones inside the ribosomal exit tunnel. Nature Struct. Mol. Biol. 12, 1123–1129 (2005).
Woolhead, C. A., McCormick, P. J. & Johnson, A. E. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell 116, 725–736 (2004).
O'Brien, E. P., Hsu, S.-T. D., Christodoulou, J., Vendruscolo, M. & Dobson, C. M. Transient tertiary structure formation within the ribosome exit port. J. Am. Chem. Soc. 132, 16928–16937 (2010).
Elcock, A. H. Molecular simulations of cotranslational protein folding: fragment stabilities, folding cooperativity, and trapping in the ribosome. PLoS Comput Biol. 2, e98 (2006).
Ferbitz, L. et al. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431, 590–596 (2004).
Kaiser, C. M. et al. Real-time observation of trigger factor function on translating ribosomes. Nature 444, 455–460 (2006).
Brandt, F. et al. The native 3D organization of bacterial polysomes. Cell 136, 261–271 (2009).
Netzer, W. J. & Hartl, F. U. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature 388, 343–349 (1997).
Frydman, J., Erdjument-Bromage, H., Tempst, P. & Hartl, F. U. Co-translational domain folding as the structural basis for the rapid de novo folding of firefly luciferase. Nature Struct. Biol. 6, 697–705 (1999).
Agashe, V. R. et al. Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed. Cell 117, 199–209 (2004).
Cuellar, J. et al. The structure of CCT–Hsc70NBD suggests a mechanism for Hsp70 delivery of substrates to the chaperonin. Nature Struct. Mol. Biol. 15, 858–864 (2008).
Zhang, G. & Ignatova, Z. Generic algorithm to predict the speed of translational elongation: implications for protein biogenesis. PLoS ONE 4, e5036 (2009).
Vabulas, R. M. & Hartl, F. U. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science 310, 1960–1963 (2005).
Buchberger, A., Bukau, B. & Sommer, T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol. Cell 40, 238–252 (2010).
Vavouri, T., Semple, J. I., Garcia-Verdugo, R. & Lehner, B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell 138, 198–208 (2009).
Arndt, V., Rogon, C. & Hohfeld, J. To be, or not to be — molecular chaperones in protein degradation. Cell. Mol. Life Sci. 64, 2525–2541 (2007).
Gamerdinger, M. et al. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 28, 889–901 (2009).
Kaganovich, D., Kopito, R. & Frydman, J. Misfolded proteins partition between two distinct quality control compartments. Nature 454, 1088–1095 (2008).
Iwata, A., Riley, B. E., Johnston, J. A. & Kopito, R. R. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 280, 40282–40292 (2005).
Kopito, R. R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524–530 (2000).
Kern, A., Ackermann, B., Clement, A. M., Duerk, H. & Behl, C. HSF1-controlled and age-associated chaperone capacity in neurons and muscle cells of C. elegans . PLoS ONE 5, e8568 (2010).
David, D. C. et al. Widespread protein aggregation as an inherent part of aging in C. elegans . PLoS Biol. 8, e1000450 (2010).
Demontis, F. & Perrimon, N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143, 813–825 (2010).
Morley, J. F. & Morimoto, R. I. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell 15, 657–664 (2004). This pioneering study provides important insight into the relationship between molecular chaperone functions and longevity.
Cohen, E., Bieschke, J., Perciavalle, R. M., Kelly, J. W. & Dillin, A. Opposing activities protect against age-onset proteotoxicity. Science 313, 1604–1610 (2006). An exciting study demonstrating that active disaggregation and the forced formation of large inclusions prevent the accumulation of toxic aggregate species in C. elegans.
Ben-Zvi, A., Miller, E. A. & Morimoto, R. I. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl Acad. Sci. USA 106, 14914–14919 (2009).
Cohen, E. et al. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell 139, 1157–1169 (2009).
Olzscha, H. et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144, 67–78 (2011). This paper demonstrates the existence of a metastable sub-proteome that is at risk of co-aggregating with amyloid-forming disease proteins.
Xu, J. et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nature Chem. Biol. 7, 285–295 (2011). This interesting study expands the range of diseases promoted by proteostasis deficiency to cancer.
Bence, N. F., Sampat, R. M. & Kopito, R. R. Impairment of the ubiquitin−proteasome system by protein aggregation. Science 292, 1552–1555 (2001). This key paper demonstrates that protein aggregation can interfere with protein degradation.
Gidalevitz, T., Ben-Zvi, A., Ho, K. H., Brignull, H. R. & Morimoto, R. I. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311, 1471–1474 (2006).
Schaffar, G. et al. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol. Cell 15, 95–105 (2004).
Lotz, G. P. et al. Hsp70 and Hsp40 functionally interact with soluble mutant huntingtin oligomers in a classic ATP-dependent reaction cycle. J. Biol. Chem. 285, 38183–38193 (2010).
Sittler, A. et al. Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington's disease. Hum. Mol. Genet. 10, 1307–1315 (2001).
Mu, T. W. et al. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell 134, 769–781 (2008).
Lee, B.-H. et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467, 179–184 (2010). This study describes the first drug-like molecule that can activate proteasome function, thus providing a means to enhance the clearance of aberrantly folded proteins.
Jahn, T. R. & Radford, S. E. The Yin and Yang of protein folding. FEBS J. 272, 5962–5970 (2005).
Vabulas, R. M., Raychaudhuri, S., Hayer-Hartl, M. & Hartl, F. U. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb. Perspect. Biol. 2, a004390 (2010).
Ryan, M. T. & Hoogenraad, N. J. Mitochondrial-nuclear communications. Annu. Rev. Biochem. 76, 701–722 (2007).
Haynes, C. M. & Ron, D. The mitochondrial UPR — protecting organelle protein homeostasis. J. Cell Sci. 123, 3849–3855 (2010).
Kenyon, C. J. The genetics of ageing. Nature 464, 504–512 (2010).
Westerheide, S. D., Anckar, J., Stevens, S. M., Jr, Sistonen, L. & Morimoto, R. I. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science 323, 1063–1066 (2009).
Acknowledgements
We thank W. Balch, A. Dillin, J. Kelly, R. Morimoto and P.Reinhart for discussions about proteostasis, and thank the members of our laboratory for comments on the manuscript. We apologize to all those whose important work could not be cited owing to space limitations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
F.U.H. is a consultant to Proteostasis Therapeutics.
Additional information
Reprints and permissions information is available at http://www.nature.com/reprints.
Rights and permissions
About this article
Cite this article
Hartl, F., Bracher, A. & Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 (2011). https://doi.org/10.1038/nature10317
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10317
This article is cited by
-
Small molecule inhibitor targeting the Hsp70-Bim protein–protein interaction in estrogen receptor-positive breast cancer overcomes tamoxifen resistance
Breast Cancer Research (2024)
-
Misfolded protein oligomers: mechanisms of formation, cytotoxic effects, and pharmacological approaches against protein misfolding diseases
Molecular Neurodegeneration (2024)
-
Nuclear Hsp104 safeguards the dormant translation machinery during quiescence
Nature Communications (2024)
-
Dynamic stability of Sgt2 enables selective and privileged client handover in a chaperone triad
Nature Communications (2024)
-
FUS maintains TAZ fluidity and function
Nature Cell Biology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.