Abstract
Megalencephaly is a congenital condition characterized by severe overdeveloped brain size. This phenotype is often caused by mutations affecting the RTK/PI3K/mTOR (receptor tyrosine kinase-phosphatidylinositol-3-kinase-AKT) signaling and its downstream pathway of mammalian target of rapamycin (mTOR). Here, using a whole-exome sequencing in a Moroccan consanguineous family, we show that a novel autosomal-recessive neurological condition characterized by megalencephaly, thick corpus callosum and severe intellectual disability is caused by a homozygous nonsense variant in the HERC1 gene. Assessment of the primary skin fibroblast from the proband revealed complete absence of the HERC1 protein. HERC1 is an ubiquitin ligase that interacts with tuberous sclerosis complex 2, an upstream negative regulator of the mTOR pathway. Our data further emphasize the role of the mTOR pathway in the regulation of brain development and the power of next-generation sequencing technique in elucidating the genetic etiology of autosomal-recessive disorders and suggest that HERC1 defect might be a novel cause of autosomal-recessive syndromic megalencephaly.
Similar content being viewed by others
Introduction
Megalencephaly is defined as an oversized and overweight brain that exceeds the age-related mean by 2 or more standard deviations and is often associated with other growth anomalies and severe intellectual disability (ID).1 Disruptions of various stages of brain development, neuronal growth, proliferation and/or migration are believed to be the underlying causes of the malformation.2 The PI3K/AKT/mTOR (phosphatidylinositol-3-kinase/AKT/mammalian target of rapamycin) pathway controls key cellular responses such as cell growth and proliferation, survival, migration and metabolism; mutations in various core members and upstream regulators of this pathway are responsible for a large proportion of megalencephaly-related disorders.2 De novo mutations in PIK3CA, AKT3 and MTOR are found in 30% of cases of hemimegalencephaly,3 whereas de novo and postzygotic mutations in AKT3, PIK3R2, PIK3CA and CCND2 cause 74% of cases of megalencephaly-capillary malformation and megalencephaly-polymicrogyria-polydactyly-hydrocephalus.4, 5 Moreover, mutations in other components of mTOR could manifest neurological symptoms without megalencephaly, such as ID, autism or epilepsy, as seen in patients with mutations in TSC1 (tuberous sclerosis complex 2), TSC2, PINK1 and DISC1.6 These findings strongly support the role of the mTOR pathway in the development and function of the brain. In this study, we provide evidence linking mutation in HERC1, another regulator of the mTOR pathway, to a distinct form of megalencephaly.
Materials and methods
Ethics statement
Institutional research ethics approval and written consent were obtained for all participants in the study.
Autozygosity mapping
Single-nucleotide polymorphism (SNP) genotyping from the proband, the two unaffected siblings and the parents was carried out on GeneChip Human Mapping 250 K Array (Affymetrix, Santa Clara, CA, USA). Linkage analysis was performed with the Merlin program using a fully penetrant autosomal-recessive inheritance model.
Exome sequencing and analyses
Exome was captured using the Agilent SureSelect Human All Exon 50 Mb Kit (Agilent Technologies, Santa Clara, CA, USA) and sequenced on a SOLiD 5500 XL machine (Life Technologies, Waltham, MA, USA). Approximately 4 Gb of sequences were produced allowing a mean sequence coverage of at least 55 reads per basepair (bp) position with >70% of captured sequences covered at 15x. Sequence reads were aligned to the human reference genome (GRCh37/HG19) using Burrow-Wheeler Aligner. Downstream processing was carried out with the Genome Analysis Toolkit (GATK),7 SAMtools8 and Picard (http://broadinstitute.github.io/picard). Variant calls were made with a GATK Unified Genotyper. Poorly mapped (with a read coverage ≤2 ×) and low-quality reads (with a Phred-scaled SNP quality of ≤20) were removed. All variants were annotated using an annotation software system that was developed in-house.
Sanger sequencing, cell culture, protein extraction and western blot
Culturing of primary skin fibroblast, protein extraction and Sanger sequencing were performed as described previously.9 Western blots were performed using conditions described previously.10
RNA extraction and reverse transcription quantitative PCR
Total RNA was extracted using Trizol Reagent (Life Technologies) and RNeasy Mini Kit (Qiagen, Venlo, Netherlands) following the manufacturer’s protocol. Reverse transcription was carried out using random hexamers and the SuperScript II Reverse Transcriptase (Life Technologies) following the manufacturer’s protocol. Quantitative PCR analysis was performed with the Power SYBR Green PCR Master Mix (Life Technologies) on an Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, Waltham, MA, USA). Specific primers were used to amplify HERC1, F-5′-TCTCCTGATTCCCAGTCAGC-3′ and R-5′-CCCTAGCACCTGTAGCTTCC-3′, and the reference gene GAPDH, F-5′-GAGTCAACGGATTTGGTCGT-3′ and R-5′-TTGATTTTGGAGGGATCTCG-3′.
Results
Clinical details
We ascertained a proband born to healthy first cousin parents of Moroccan origin who also have three healthy children. He was born at term after a normal pregnancy, and growth parameters at birth were: weight 4000 g (90th percentile), length 53 cm (90th percentile) and OFC 37 cm (90th percentile). In the first years of life, height (85, 98, 122, 137, 153 at 2, 4, 8, 11 and 14 years of age, respectively) and weight (12, 15, 23, 28, 37 at 2, 4, 8, 11 and 14 years of age, respectively) curves progressively declined at −1 SD but the OFC curve was above +3 SD, with a final OFC at 66.5 cm at 18 years of age, contrasting with normal parameters for weight (51 kg, −1 SD) and height (172 cm, M). Skeleton X-rays were normal with no advanced bone age or scoliosis and he had a normal puberty at 13 years of age. He had severe developmental delay with no speech, limited social interaction, major difficulties with walking (first steps at 5 years of age) with joint limitations and sleep disturbances with waking up early. At 4 years of age, he developed generalized epilepsy, which was well controlled by a combination of carbamazepine and clobazam. Facial features included hypotonic and long face with high forehead, prognatism, salivary incontinence and long and thin feet and hand. He also presented severe myopia, detected at 5 years of age, and repeated pulmonary infections. Brain MRI demonstrated bilateral megalencephaly, a thick corpus callosum, enlarged white matter, septum pellucidum cyst and a small cerebellum (Figure 1b). The phenotype of the patients has been uploaded to the MedGen Database (NCBI) under the accession number 832891.
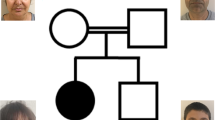
A truncation variant in HERC1 is associated with a distinct form of megalencephaly syndrome. (a) Family pedigree. Black box indicates the affected proband. White box with small black dot indicates carrier. (b) Magnetic resonance imaging (MRI) of the affected proband demonstrating bilateral megalencephaly, a thick corpus callosum, enlarged white matter, septum pellucidum cyst and a small cerebellum. (c) Autozygosity mapping results. Y axis represents LOD (logarithm (base 10) of odds) score, X axis indicates the chromosomes, red bars indicate the position of the homozygous region presenting in the affected proband and black arrow indicates the position of the HERC1 gene. Owing to small family size, no IBD region reaches the significant LOD threshold of at least 2. (d) Variant validation and segregation testing. Sanger sequencing was used to validate the variant identified by WES. The proband is homozygous and other family members are heterozygous for the c.9748C>T variant.
Genetic investigations
To investigate the genetic etiology, we first performed multipoint linkage analysis in the affected proband, the two healthy siblings and the parents. Large blocks of homozygous identical-by-descent (IBD) regions were identified; however, none reached the significant LOD score threshold (Figure 1c). We then performed WES using DNA extracted from the blood of the proband and the parents. Alignment and variant calling were performed as described previously.9 Variants previously reported in the databases dbSNP, 1000Genomes and Exome Variant Server with minor allele frequency >1% were excluded. Variants were also filtered against an in-house database (>2000 exomes), and only nonsense, missense, insertions/deletions and variants affecting splice sites were further studied. Two homozygous nonsense variants in the genes EIF2S3L (NM_001415.3:c.220G>A (p.Glu74*)) and HERC1 (NM_003922.3:c.9748C>T (p.Arg3250*)) fulfilled these criteria under a recessive inheritance model. Filtering WES data for compound heterozygous variants, X-linked or heterozygous 'de novo' variant did not provide any additional candidate variant. Both variants were confirmed by capillary sequencing. In the case of the EIF2S3L variant, both parents are heterozygous for the variant, whereas it is in homozygous form in the unaffected sister (data not shown). Moreover, this variant has been reported 12 times (including once in homozygous form) in 60 706 unrelated individuals from The Exome Aggregation Consortium (ExAC) corresponding to an allele frequency of 9.893e−05. Thus, this variant is unlikely to be responsible for the phenotype observed. By contrast, the c.9748C>T HERC1 variant is absent from all publicly available data set. Capillary sequencing showed that the parents and both healthy siblings are heterozygous for this variant (Figure 1d); the youngest healthy brother (individual II.4) could not be tested owing to unavailability of the DNA. HERC1 is among the genes most intolerant to functional variation with a low residual variation intolerance score (−3.88; 0.21%). Lastly, this variant lies within a 10.9-Mb-long IBD region on chromosome 15 (Figure 1c). Collectively, these data support the pathogenicity of the c.9748C>T HERC1 variant and its link to the phenotype observed in our patient. It is now annotated in the ClinVar Database (NCBI) under the accession number SCV000212106. Three other patients in our cohort with a similar phenotype were screened for variants in HERC1 using WES but no additional variant was identified.
Absence of the HERC1 protein in patient cells
HERC1 is a 78-exon-long gene spanning 225 kb on chr15q22.31. It encodes for a large and highly conserved ubitiquin E3 ligase protein (>97% amino-acid identity in mouse). The homozygous c.9748C>T variant lies within exon 49, and is expected to cause premature termination of translation. The impact of the variant on HERC1 expression was first assessed by reverse transcription quantitative PCR (RT-qPCR) of cultured skin fibroblast RNA, which indicated that HERC1 mRNA level is at least 50 times less than the controls (Figure 2a). The total absence of the full-length and the potential truncated product was confirmed by immunoblot analysis of primary patient skin fibroblast lysates using two different anti-HERC1 antibodies, Bvg5 and 410, which target the N or C terminus of the protein, respectively (Figure 2b). These results suggest that the identified variant leads to nonsense-mediated decay of HERC1 mRNA and that the phenotype of the proband is likely manifested by the complete loss of function of HERC1 protein.
The c.9748C>T, p.R3250* variant leads to complete loss of HERC1 protein. (a) Reverse transcription quantitative PCR (RT-qPCR) analysis showing the expression of HERC1 mRNA (±SD) in the proband and two unrelated controls. Comparative Ct method was used to calculate gene expression, and GAPDH was used as reference gene. (b) Top panel, schematic showing the areas targeted by the two different anti-HERC1 antibodies Bgv5 and 410. Bottom panel, western blot showing the complete loss of HERC1 protein in the proband. No change was observed for HERC2, mTORC1 and TSC2. α-Tubulin was used as the loading control.
The C-terminus region of HERC1 was shown to interact directly with TSC2, preventing the formation of a functional TSC with TSC1, which represses the mTORC1 pathway;10 these authors also suggested a possible model where HERC1 may target TSC2 for degradation. Thus, we tested the level of TSC2 and the mTORC1 complex in patient cells by western blot, but we did not observe any change in either TSC2 or mTORC1 protein levels (Figure 2b). Lastly, we tested for the functional compensation of HERC1 absence by its closely related paralog HERC2; we investigated the level of HERC2 protein by western blot analysis in patient cells. No obvious difference in HERC2 protein level was observed (Figure 2b), suggesting that HERC2 does not over compensate for the loss of HERC1.
Discussion
We identified a loss-of-function homozygous variant in HERC1 in a proband with a novel form of megalencephaly. Disorders due to HERC1 variants are likely to be rare. In our cohort of about 100 consanguineous families, we did not identify any other patient linked to this locus. Moreover, in the literature, the association of megalencephaly and thick corpus callosum has only been previously reported in three unrelated children who also had pachygyria and complete lack of motor development, features that are not observed in our case.11 Conversely, cerebellar atrophy was not reported in these patients.
The HERC1 protein belongs to the HERC protein family of ubiquitin ligases characterized by the presence of an HECT domain and one or more RCC1-like domains.12 This family consists of two subgroups according to their sizes and domain architecture. The four small proteins (HERC3–6) possess little more than the two above-mentioned domains. The two giant HERC1 and HERC2 proteins also contain other functional domains, including two RLDs, a C-terminal HECT, a WD40, a SPRY (spl A and RyR) domain and several other minor motifs.12
In zebrafish, the HERC1 orthologous gene (LOC569603) is not expressed in the brain suggesting functional divergence between the human and the zebrafish protein and preventing the use of this model to understand how loss of HERC1 protein alter brain development and function (Supplementary Information). By contrast, the murine HERC1 gene is ubiquitously expressed and a variety of mutations have been reported in several members of the HERC families, leading to sterility, growth retardation, retinitis pigmentosa, amyotrophic lateral sclerosis or cancer.13, 14 Notably, the tambaleante (tbl) mutant mice is caused by a homozygous missense mutation (p.Gly483Glu) in HERC1, resulting in progressive Purkinje cell degeneration, severe ataxia and growth retardation.15 In contrast to what is observed in our patient, the Herc1tbl mutation enhances the stability of the Herc1 protein. However, the observation of cerebellar atrophy in our patient confirms the role of this protein in the growth and maintenance of the cerebellar structure. In human, loss-of-function mutations in HERC2 lead to global developmental delay, autism with Angelman-like features.9, 10 The identification of a HERC1 variant in a novel neurological condition further supports the importance of the large HERC protein family in brain development.
HERC1 protein interacts with TSC2 and negatively regulates the mTORC1 pathway.10 Although we did not observe any significant change in the expression level of TSC2 or mTORC1 in skin fibroblast of the patient (Figure 2b), we cannot discard that the involvement of HERC1 in the mTOR pathway could be tissue-specific. Given that the megalencephaly phenotype is largely attributed to misregulation of the mTOR pathway,3, 4, 5 it is likely that loss of HERC1 could impair this pathway early in brain development.
Loss of HERC1 protein may also affect intracellular membrane trafficking. Indeed, the N-terminus RLD1 domain acts as a GEF guanine nucleotide exchange factor (GEF) for small GTPases and can stimulate guanine nucleotide dissociation from ARF1 and ARF6 GTPases.12 The ARF proteins are master mediators of membrane trafficking and vesicular transport;16 misregulation of the ARF proteins causes not only lysosomal-related disorders17 but also ID as seen in IQSEC2 patients in whom loss of IQSEC2 protein impairs ARF6 activation.18 It is thus likely that the pathological mechanism underlying the loss of HERC1 involves several pathways, all of which have significant roles in early brain development and function.
In summary, we report here a homozygous loss-of-function variant in HERC1 leading to complete loss of the protein in a proband with the unusual association of megalencephaly, thick corpus callosum, cerebellar atrophy and severe ID. Although the final demonstration of our hypothesis is awaiting the identification of additional HERC1 variants in other patients with similar phenotypes, these results make HERC1 an excellent candidate gene for megalencephaly. Lastly, our result emphasizes the roles of the mTOR pathway and its regulatory effectors in the development of the brain, and calls for further investigation of the role of HERC1 in early neurogenesis in the cortex and the cerebellum.
References
Olney AH : Macrocephaly syndromes. Semin Pediatr Neurol 2007; 14: 128–135.
Mirzaa GM, Poduri A : Megalencephaly and hemimegalencephaly: breakthroughs in molecular etiology. Am J Med Genet C 2014; 166C: 156–172.
Lee JH, Huynh M, Silhavy JL et al: De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet 2012; 44: 941–945.
Riviere JB, Mirzaa GM, O'Roak BJ et al: De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet 2012; 44: 934–940.
Mirzaa GM, Parry DA, Fry AE et al: De novo CCND2 mutations leading to stabilization of cyclin D2 cause megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome. Nat Genet 2014; 46: 510–515.
Lipton JO, Sahin M : The neurology of mTOR. Neuron 2014; 84: 275–291.
McKenna A, Hanna M, Banks E et al: The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303.
Li H, Handsaker B, Wysoker A et al: The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–2079.
Langouet M, Saadi A, Rieunier G et al: Mutation in TTI2 reveals a role for triple T complex in human brain development. Hum Mutat 2013; 34: 1472–1476.
Chong-Kopera H, Inoki K, Li Y et al: TSC1 stabilizes TSC2 by inhibiting the interaction between TSC2 and the HERC1 ubiquitin ligase. J Biol Chem 2006; 281: 8313–8316.
Gohlich-Ratmann G, Baethmann M, Lorenz P et al: Megalencephaly, mega corpus callosum, and complete lack of motor development: a previously undescribed syndrome. Am J Med Genet 1998; 79: 161–167.
Garcia-Gonzalo FR, Rosa JL : The HERC proteins: functional and evolutionary insights. Cell Mol Life Sci 2005; 62: 1826–1838.
Ji Y, Walkowicz MJ, Buiting K et al: The ancestral gene for transcribed, low-copy repeats in the Prader–Willi/Angelman region encodes a large protein implicated in protein trafficking, which is deficient in mice with neuromuscular and spermiogenic abnormalities. Hum Mol Genet 1999; 8: 533–542.
Lehman AL, Nakatsu Y, Ching A et al: A very large protein with diverse functional motifs is deficient in rjs (runty, jerky, sterile) mice. Proc Natl Acad Sci USA 1998; 95: 9436–9441.
Mashimo T, Hadjebi O, Amair-Pinedo F et al: Progressive Purkinje cell degeneration in tambaleante mutant mice is a consequence of a missense mutation in HERC1 E3 ubiquitin ligase. PLoS Genet 2009; 5: e1000784.
Donaldson JG, Jackson CL : ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol 2011; 12: 362–375.
Seixas E, Barros M, Seabra MC, Barral DC : Rab and Arf proteins in genetic diseases. Traffic 2013; 14: 871–885.
Shoubridge C, Tarpey PS, Abidi F et al: Mutations in the guanine nucleotide exchange factor gene IQSEC2 cause nonsyndromic intellectual disability. Nat Genet 2010; 42: 486–488.
Acknowledgements
We thank the family for the participation in this study. We acknowledge the technical contribution of the Genomic and Bioinformatic Platforms of the Institute Imagine, Hôpital Necker-enfants malades. We acknowledge the use of the bioresources of the Necker Imagine DNA biobank (BB-033-00065). This program has received a state subsidy managed by the National Research Agency under the ‘Investments for the Future’ program bearing the reference ANR-10-IAHU-01. This study was also supported by the Centre National de la Recherche Scientifique (CNRS), the Fondation pour la Recherche Médicale (DEQ20120323702) and the Ministère de la Recherche et de l′Enseignement Supérieur (to LC), and by the Spanish Ministerio de Ciencia e Innovación (BFU2011-22498) (to JLR). TS was supported by a fellowship from CAPES Foundation, Ministry of Education of Brazil.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Nguyen, L., Schneider, T., Rio, M. et al. A nonsense variant in HERC1 is associated with intellectual disability, megalencephaly, thick corpus callosum and cerebellar atrophy. Eur J Hum Genet 24, 455–458 (2016). https://doi.org/10.1038/ejhg.2015.140
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2015.140
This article is cited by
-
Mutation of the HERC 1 Ubiquitin Ligase Impairs Associative Learning in the Lateral Amygdala
Molecular Neurobiology (2018)
-
Complete loss of function of the ubiquitin ligase HERC2 causes a severe neurodevelopmental phenotype
European Journal of Human Genetics (2017)