-
PDF
- Split View
-
Views
-
Cite
Cite
Zhenzhong Li, Youyi Zhang, Li Ku, Keith D. Wilkinson, Stephen T. Warren, Yue Feng, The fragile X mental retardation protein inhibits translation via interacting with mRNA, Nucleic Acids Research, Volume 29, Issue 11, 1 June 2001, Pages 2276–2283, https://doi.org/10.1093/nar/29.11.2276
Close - Share Icon Share
Abstract
Fragile X syndrome is a frequent form of inherited mental retardation caused by functional loss of the fragile X mental retardation protein, FMRP. The function of FMRP is unknown, as is the mechanism by which its loss leads to cognitive deficits. Recent studies have determined that FMRP is a selective RNA-binding protein associated with polyribosomes, leading to the hypothesis that FMRP may be involved in translational regulation. Here we show that purified recombinant FMRP causes a dose-dependent translational inhibition of brain poly(A) RNA in rabbit reticulocyte lysate without accelerated mRNA degradation. In our translation reaction FMRP interacts with other messenger ribonucleoproteins and pre-exposure of FMRP to mRNA significantly increased the potency of FMRP as a translation inhibitor. Translation suppression by FMRP is reversed in a trans-acting manner by the 3′-untranslated portion of the Fmr1 message, which binds FMRP, suggesting that FMRP inhibits translation via interacting with mRNA. Consistently FMRP suppresses translation of the parathyroid hormone transcript, which binds FMRP, but not the β-globin transcript, which does not bind FMRP. Moreover, removing the FMRP-binding site on a translation template abolishes the inhibitory effect of FMRP. Taken together, our results support the hypothesis that FMRP inhibits translation via interactions with the translation template.
Received February 28, 2001; Revised and Accepted April 6, 2001.
INTRODUCTION
Absence of the protein encoded by the FMR1 gene leads to fragile X syndrome, a frequent cause of familial mental retardation (1–4). This protein, designated the fragile X mental retardation protein (FMRP), harbors RNA-binding motifs, including two K homology (KH) domains as well as an RGG box, and has been demonstrated to bind RNA in a selective manner (5–7). The RNA-binding activity of FMRP appears to be an intrinsic property of this protein, since purified recombinant FMRP also binds RNA in vitro with some RNA selectivity (7). Thus, it is generally believed that the function of FMRP is closely associated with its RNA-binding activities. Indeed, FMRP is incorporated into cellular messenger ribonucleoprotein (mRNP) particles (8,9). These mRNP particles associate with large polyribosomal complexes in the cytoplasm of various cell types (8–10), including those in the somatodendritic compartments of brain neurons (11). Within the FMRP-containing polyribosomal mRNP particles FMRP appears to interact with several other proteins, including its autosomal homologs FXR1P and FXR2P (12) and nucleolin (13), as well as a novel RNA-binding nuclear protein NUFIP (14). Besides the presence of RNA-binding motifs, FMRP also carries a nuclear localization and a nuclear export signal (NLS and NES) and presumably shuttles between the nucleus and the cytoplasm (11,15,16).
The association of FMRP with the translation machinery as a mRNP component has been studied by a number of laboratories (8–11,17), leading to the hypothesis that mRNA binding by FMRP may be involved in translational regulation. To test this hypothesis we have examined the effect of recombinant FMRP on translation in rabbit reticulocyte lysate (RRL), which is commonly used to demonstrate the influence on translation of many other RNA-binding proteins (18–22). We report here that purified recombinant FMRP can suppress translation of brain poly(A) RNA in a dose-dependent manner. This is not due to a general impairment of the translation machinery, since high levels of FMRP did not cause detectable changes in translation of a poly(A) RNA pool composed largely of globin mRNA. We also show that recombinant FMRP interacts with endogenous mRNPs in the RRL, including FXR2P, a known in vivo FMRP partner. Pre-incubating FMRP with translation template mRNAs increased the potency of FMRP as a translation inhibitor. In addition, the 3′-untranslated region (3′‐UTR) of the Fmr1 transcript, which has been reported to bind FMRP in vitro (7), reversed the translation inhibition caused by FMRP in a trans-acting manner, suggesting that the inhibitory effect of FMRP on translation is mediated by interactions between FMRP and the template mRNA. Consistent with this notion, transcripts that do not bind FMRP are insensitive to translation suppression by FMRP. Furthermore, removing the FMRP-binding sequence of an mRNA abolishes the inhibitory effect of FMRP. These data support the hypothesis that FMRP may function as a negative regulator for translation and interaction with the mRNA is essential for FMRP to inhibit translation.
MATERIALS AND METHODS
Preparation of translation templates
Cerebral cortical total RNA was isolated from 2–3 month old male Sprague–Dawley rats by Trizol extraction (Life Technologies, Gaithersburg, MD). Using the same method, total RNA was also isolated from rabbit reticulocyte-rich whole blood (Pel-Freez, Rogers, AR). Poly(A) RNA was further purified from these total RNA preparations using an Oligo™ mRNA isolation kit (Qiagen, Valencia, CA). The enrichment of poly(A) RNA was confirmed by visualization of the RNAs on ethidium bromide stained agarose gels. The quantity of poly(A) RNA was determined from the OD260. The BamHI–XhoI fragment containing the entire 3′-UTR of the fmr1 cDNA (23) was subcloned into Bluescript SK (Stratagene, La Jolla, CA) to generate the plasmid ΔBam. The transcript containing the murine fmr1 3′-UTR was prepared by in vitro transcription using XhoI-linearized ΔBam. The rabbit β-globin cDNA was generated from a RT–PCR reaction using rabbit reticulocyte poly(A) RNA with primers 5′-ACACTTGCTTTTGACACAAC-3′ and 5′-AGCCAGAAGTCAGATGCTC-3′ (24) and cloned into Bluescript SKSII. Two clones with opposite cDNA orientations were used to generate the full-length sense strand and the antisense strand of the β-globin 3′-UTR with T7 polymerase. The full-length sense strand was generated from XhoI-linearized plasmid. The antisense transcript was generated using the plasmid linearized at the NcoI internal site of the β-globin cDNA. The capped parathyroid hormone (PTH) mRNA template was generated with T3 polymerase using PstI-linearized plasmid (25) and the mCAP RNA Capping kit (Stratagene). The capped full-length and the 3′‐UTR-truncated transcript encoding the 18 kDa myelin basic protein (MBP) was generated with T7 RNA polymerase using BamHI-linearized cDNA constructs generously provided by Dr Campagnoni (UCLA, Los Angeles, CA) (26).
Recombinant FMRP
Purification of recombinant Flag–FMRP from a baculovirus expression system was as previously described (7). Briefly, SF9 cells were infected by the baculovirus construct encoding Flag-tagged full-length FMRP. Native recombinant Flag–FMRP was purified by binding to an anti-Flag M2 affinity column and elution by competition with Flag–peptide, followed by dialysis. The Bradford assay (Bio-Rad, Hercules, CA) was used to estimate the quantity of Flag–FMRP (7). The purity of Flag–FMRP was confirmed by Coomassie staining of SDS–PAGE gels and the amount of Flag–FMRP used in each translation reaction was determined by normalizing the intensity of Coomassie stained Flag–FMRP to known amounts of BSA (New England Biolabs, Beverly, MA).
Northern hybridization
After incubation with FMRP or mock treatment total RNA was isolated before and after exposure to RRL by Trizol extraction or phenol/chloroform extraction. RNA samples were then subjected to northern hybridization analysis as previously described (27). 32P-labeled probes were generated by random labeling (Amersham Pharmacia, Piscataway, NJ) using purified cDNA fragments, including those encoding rat glyceraldehyde phosphate dehydrogenase (GAPDH) (27), ribosomal protein S14 (RPS14) (27), MBP (26) and 28S ribosomal RNA (28).
Immunoprecipitation and immunoblot analysis
Immunoprecipitation of FMRP from the translation mix was performed using an anti-Flag M2 affinity gel (Sigma, St Louis, MO) following the manufacturer’s protocol. The bound proteins were eluted by heating at 95°C in SDS sample buffer for 3 min and subjected to SDS–PAGE immunoblot analysis. Immunoblot analysis was carried out as previously described (27). The anti-Flag M2 monoclonal antibody (Sigma) was used to detect FMRP and A42 monoclonal antibody was used to detect FXR2P (12).
In vitro translation reaction
For each translation reaction 1.5 µg mRNA template was heated at 65°C for 3 min before addition of 20 µCi [35S]methionine (Amersham Pharmacia) and 10 U RNase block (Stratagene) on ice. The mixture was then exposed to various amounts of FMRP in 5 µl of PBS containing 250 mM NaCl and incubated on ice for 3 min before initiation of translation by addition of 40 µl of RRL (Stratagene). Incubation of translation template mRNA with the same buffer containing no FMRP was defined as mock treatment. The final concentration of recombinant FMRP in the translation reaction was from 0 to 250 nM. An aliquot of each translation reaction was subjected to trichloroacetic acid (TCA) precipitation at 10 min after initiation following the manufacturer’s protocol (Stratagene). The TCA precipitable radioactivity (c.p.m.) was measured by scintillation counting. Parallel translation reactions containing no mRNA were performed to determine the background. Translation products were also subjected to SDS–PAGE, followed by phosphorimager analysis.
Quantitative and statistical analyses
Background counts were subtracted from the TCA precipitable c.p.m. of each translation reaction and the adjusted c.p.m. derived from the mock-treated reaction was defined as 100%. For individual mRNA templates the translation level was determined by phosphorimager quantification of the 35S‐labeled peptide on SDS–PAGE gels. Again, the mock-treated reaction was defined as 100%. TCA precipitable c.p.m. or phosphorimager units derived from each FMRP-treated reaction were normalized to that from the mock-treated reaction and used to calculate translation yield. The result was subjected to one-way ANOVA or paired t-tests as indicated in the corresponding figure legends and the P value for each experiment was calculated based on Newman–Keuls multiple comparison tests.
RNA binding assay
[35S]Methionine-labeled FMRP was generated by the TNT reaction (Promega, Madison, WI) and then incubated with biotinylated RNA synthesized by in vitro transcription (Stratagene). The biotinylated PTH and β-globin transcripts were generated with T3 and T7 RNA polymerase from linearized plasmid as described above. The biotinylated MBP transcripts were generated with T7 RNA polymerase. RNA binding reactions were carried out and the bound [35S]FMRP was captured on streptavidin-conjugated Dynabeads as described previously (5). The captured [35S]FMRP was fractionated by SDS–PAGE, followed by phosphorimager analysis, or subjected to scintillation counting.
RESULTS
FMRP causes a dose-dependent inhibition of the translation of brain poly(A) RNA but not of reticulocyte poly(A) RNA
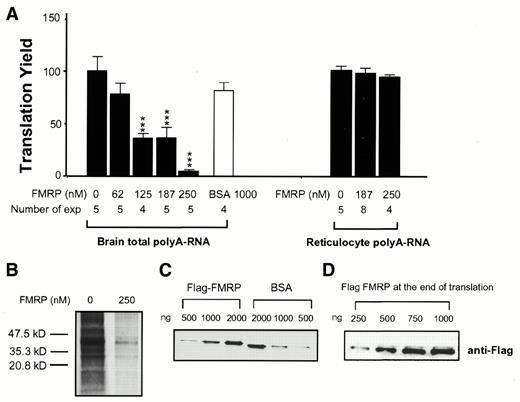
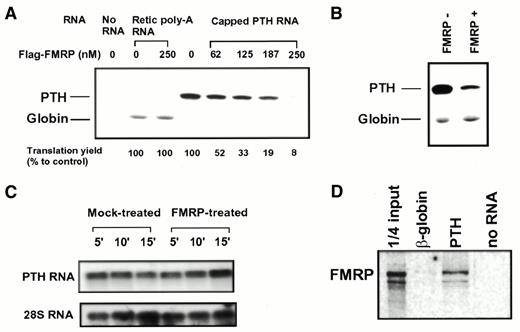
One common way to directly address the influence of an RNA-binding protein on translation is to examine its effect in a cell-free in vitro translation system (18,20). Since the physiological mRNA target(s) for FMRP has yet to be defined, we first chose to examine the influence of FMRP on translation using total rat brain cortical poly(A) RNA as translation templates in the RRL translation system. The translation yield for each reaction was estimated by the incorporation of [35S]methionine into the newly synthesized peptide as measured by the TCA precipitable radioactivity (c.p.m.). In order to allow time for FMRP to interact with the mRNA, poly(A) RNA was pre-incubated with FMRP before translation took place. The TCA precipitable c.p.m. in the mock-treated reaction, in which mRNA was pre-incubated with buffer lacking FMRP, was defined as 100%. As shown in Figure 1A, a reduction of >50% in the total translation yield was observed in the presence of 125 nM FMRP at 10 min after translation initiation (P < 0.001). This concentration of FMRP corresponds to a molar ratio of FMRP to translation templates of ∼1:1. Increasing amounts of FMRP resulted in a further reduction in the translation yield. SDS–PAGE analysis indicated that pre-incubating translation templates with a high concentration of FMRP can lead to translational inhibition of a rather broad spectrum of mRNA species (Fig. 1B). In contrast, much higher levels of BSA did not influence translation significantly in parallel experiments. A similar inhibitory effect of FMRP was also observed when rat liver poly(A) RNA was used as translation template (data not shown). However, a high dose of FMRP did not affect translation of poly(A) RNA isolated from rabbit reticulocytes, in which the majority of the poly(A) RNA encodes globin. This result argues that the effect of FMRP is unlikely to be due to a general impairment of the translation machinery, but is determined by the mRNA species. The quantity of Flag–FMRP added in each translation reaction was verified by SDS–PAGE followed by Coomassie staining using BSA as the standard (Fig. 1C). The integrity of FMRP during translation was confirmed by immunoblotting of the reaction mix containing various amounts of recombinant FMRP at the end of incubation using anti-Flag antibody (Fig. 1D).
Inhibition of translation by FMRP does not accelerate RNA degradation
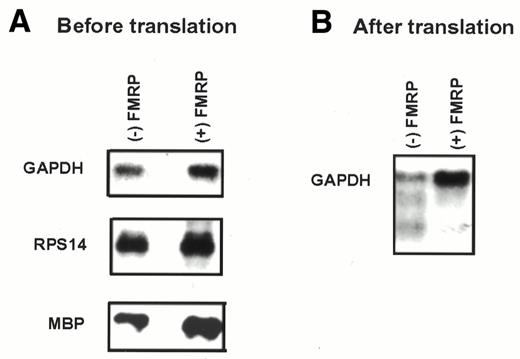
To eliminate the possibility that the reduced translation yield caused by FMRP may result from degradation of the translation templates we isolated RNA with or without exposure to FMRP and examined the quantity of various transcripts by northern hybridization analysis. As shown in Figure 2A, exposure to FMRP before translation did not result in detectable degradation of mRNA. Furthermore, when comparing the mRNA level at the end of translation the level of translation templates recovered from the FMRP-treated reaction was not reduced in comparison to those recovered from the mock-treated reaction (Fig. 2B). In fact, the presence of FMRP appears to protect the mRNA from degradation to a certain degree. Therefore, inhibition of protein synthesis by FMRP in our system is unlikely to be due to translation-coupled degradation of the mRNA templates, but instead appears to be due to inhibition of translation.
FMRP interacts with other mRNPs in RRL and the potency of FMRP as a translation inhibitor is enhanced by pre-exposure to the translation template
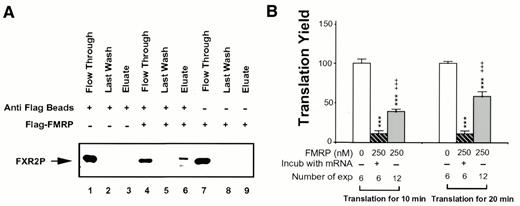
FMRP has been shown to interact with other mRNPs in vivo, including its autosomal homologs FXR1P and FXR2P (12,22). To address whether exogenously added recombinant FMRP may form similar interactions with endogenous mRNPs in RRL we immunoprecipitated recombinant FMRP from the translation reaction mix using an anti-Flag M2 affinity gel. As shown in Figure 3A, FXR2P was co-immunoprecipitated with FMRP (lane 6). This result suggested that recombinant FMRP indeed associated with other mRNPs in our translation system. Not all the FXR2P in RRL could be co-precipitated with the recombinant FMRP (Fig. 3A, lane 4). This may suggest pre-occupation of the FXR2P by endogenous FXRP, FMRP and/or other mRNPs. The interaction between FMRP and FXR2P in RRL did not require the presence of template mRNA (data not shown). We also compared the effect of FMRP on translation with or without pre-incubating FMRP with the translation template mRNAs. As shown in Figure 3B, pre-incubation of FMRP with brain poly(A) RNA significantly increased the potency for FMRP as a translation inhibitor. This result suggests that translational inhibition by FMRP may be quantitatively modulated by interactions of FMRP with other mRNPs prior to the association of FMRP with the mRNA.
The FMRP–mRNA interaction is critical for FMRP to inhibit translation
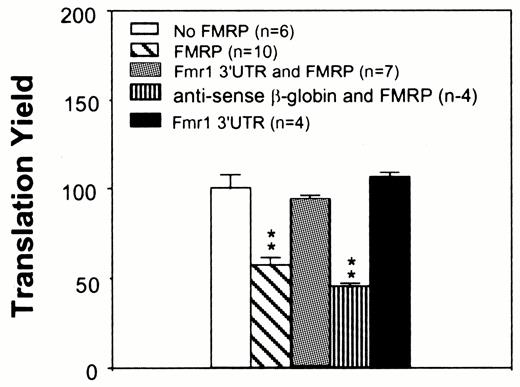
If the FMRP–mRNA interaction is required for FMRP to exert its inhibitory effect on translation the presence of a competitor RNA with high affinity for FMRP may reverse the effect of FMRP on other translation templates. Indeed, the presence of the Fmr1 3′-UTR, which has been shown to bind FMRP in vitro (7), reversed the inhibitory effect of FMRP on translation of brain poly(A) RNA (Fig. 4). Such reversal was unlikely to be due to peptide production from the Fmr1 3′-UTR, since it yielded only negligible peptide as measured by [35S]methionine incorporation by SDS–PAGE (data not shown). In contrast, the antisense strand of the β-globin 3′-UTR did not bind FMRP and was incapable of reversing the inhibitory effect of FMRP, suggesting that the reversal by the Fmr1 3′-UTR is most likely to be due to competitive inhibition of FMRP binding to the translation template mRNAs.
Because a large amount of FMRP did not suppress translation of rabbit reticulocyte poly(A) RNA (Fig. 1A) we tested whether FMRP may differentially affect translation of various mRNAs, presumably due to differences in the ability of the mRNA to interact with FMRP. Quantitative analysis of 35S‐labeled globin by SDS–PAGE confirmed the lack of influence of FMRP on globin synthesis (Fig. 5A, lanes 2 and 3). To test whether the lack of inhibition of globin translation by FMRP is due to the high translation efficiency of this message we examined the influence of FMRP on translation of the PTH transcript. This transcript is the standard control translation template recommended by the manufacturer and displays at least comparable translation efficiency to that of globin messages. In contrast to globin, the yield of PTH was reduced upon FMRP treatment in a dose-dependent manner (Fig. 5A, lanes 4–8), indicating that translation of the PTH message, despite being a high efficiency translation template, is suppressed by FMRP. Furthermore, we confirmed that FMRP selectively inhibits translation of the PTH message but not the globin message, even when present in the same reaction (Fig. 5B). Northern hybridization analysis confirmed that FMRP did not cause degradation of the PTH transcripts (Fig. 5C), indicating that reduced PTH production was due to translational inhibition. Using a standard RNA binding assay we further demonstrated that FMRP binds the PTH transcript in vitro, whereas only minimal FMRP binding was detected for the β-globin transcript (Fig. 5D). This result suggests that the incapability of interacting with FMRP of the β-globin transcript may explain why this transcript is insensitive to the effect of FMRP on translation.
If the interaction between FMRP and the translation template is essential for FMRP to inhibit translation one would expect that removing the FMRP-binding site on the mRNA should abolish the translational suppression caused by FMRP. Our previous work has indicated that FMRP can bind to the 3′-UTR of the MBP transcript (7). As shown in Figure 6A, we found that removing the 3′-UTR almost completely abolished the FMRP binding activity of the MBP transcript. As expected, translation of the full-length MBP transcript was inhibited by FMRP (Fig. 6B). In contrast, at a comparable molar ratio of FMRP to RNA the MBP transcript lacking the 3′-UTR fails to respond to the translation suppression caused by FMRP. This result supports the hypothesis that interaction with the translation template is required for FMRP to exert its inhibitory influence on translation.
DISCUSSION
The absence of functional FMRP leads to fragile X syndrome, which most often manifests as moderate mental retardation and other, more subtle, somatic features. However, the physiological function of FMRP as well as the consequence of lacking FMRP is poorly understood. Previous studies have demonstrated FMRP to be an RNA-binding protein associated with polyribosomes, suggesting a possible role for FMRP in influencing protein translation. A recent report by Laggerbauer et al. (29) provided evidence that bacteria-derived recombinant FMRP can inhibit translation of in vitro transcribed RNA in RRL and in injected Xenopus oocytes. The bacteria-derived recombinant FMRP was shown to inhibit translation of transcripts derived from several genes that contained only the coding sequence. Thus, although FMRP has been show to interact with the 3′-UTR of various mRNAs with high affinity (7), the importance of such an interaction between FMRP and the translation template has not been established. Whether FMRP can suppress translation of isolated cellular poly(A) RNA remains to be tested. Furthermore, whether FMRP inhibits translation of all RNA or, alternatively, FMRP only inhibits translation of its bound mRNA needs to be addressed. In this study we used recombinant FMRP generated from a baculovirus expression system and our results indicate that FMRP can suppress translation of a variety of mRNA species, including poly(A) RNA isolated from brain. Furthermore, our observations suggest that interaction with the translation template mRNA is essential for FMRP to exert its inhibitory influence on translation.
In our translation reactions the concentrations of exogenously added recombinant FMRP are estimated to range from 0.2 to 0.8 ng/µg total protein, resulting in an ∼1–4-fold excess in comparison to the endogenous FMRP level in RRL. However, it is possible that only a portion of the recombinant FMRP harbors activity for RNP complex formation and translation inhibition. Nonetheless, the FMRP levels in human peripheral blood are estimated to be ∼1.3 ng/µg total protein (A.Kenneson and S.T.Warren, manuscript in preparation). Therefore, the concentrations of FMRP in our reactions are not widely exaggerated in comparison to the physiological levels. The lowest amount of FMRP applied to brain poly(A) RNA only reduced translation by ∼20% (Fig. 1). We would propose that at low concentrations of FMRP it is likely that only certain messages, perhaps those with the highest affinity for FMRP, may be affected. At high levels of exogenous FMRP, translation yield for most messages could be reduced by >85%, suggesting that messages with low or non-specific affinity are being affected. Indeed, it has been shown previously that the RGG RNA-binding domain in FMRP can interact with RNA in vitro in a non-specific manner (30). Yet, one cannot rule out that such concentration-dependent and low affinity interactions are functional in vivo. The stoichiometry of FMRP to its mRNA target may be physiologically important, considering the divergent levels of FMRP in different cell types (31–34), during tissue remodeling (33), at different stages of the cell cycle (34) and in different subcellular compartments, such as the neuronal soma versus the dendritic spine (11). This idea is consistent with the recent finding that not only FMRP deficiency but also overexpression of FMRP in mice causes behavioral abnormalities (35).
Despite the fact that FMRP can inhibit translation of a broad spectrum of mRNA species in RRL, FMRP does not cause a general impairment of the translation machinery. This is indicated by the fact that translation of certain mRNA species is not inhibited by FMRP (Figs 5 and 6). The insensitivity of the globin message to translation inhibition by FMRP most likely results from its inability to bind to FMRP. This hypothesis is further supported by the fact that removing the FMRP-binding site on the translation template abolished the inhibitory effect of FMRP. Furthermore, the inhibitory effect of FMRP on translation was partially reversed by the Fmr1 3′-UTR in a trans-acting manner (Fig. 4), presumably due to sequestration of FMRP from interacting with the translation template mRNA. Taken together, these results support the hypothesis that FMRP inhibits translation via interacting with the mRNA.
A lengthy poly(A) tail of the translation template does not appear to be essential for the inhibitory effect of FMRP on translation. This is indicated by the fact that the PTH transcript, which showed comparable sensitivity to the effect of FMRP as that observed for brain poly(A) RNA, contains only 13 A residues in the tail (25), whereas the poly(A) tail of mammalian cellular poly(A) RNA usually contains more than 200 A residues (36).
In most of our reactions FMRP was pre-incubated with the translation templates before the initiation of translation. Such a treatment enhanced the potency of FMRP in suppressing translation (Fig. 3B), yet may reduce the RNA selectivity of the influence of FMRP. In eukaryotic cells primary transcripts are normally bound by other RNPs. Therefore, the broad spectrum of translation inhibition by pre-exposing FMRP to naked mRNA may not mimic the in vivo situation. Association of FMRP with other mRNPs may alter the activity of FMRP in interacting with mRNA, which could modulate the potency of FMRP as a translation inhibitor. This notion is supported by the fact that the inhibitory effect of FMRP on translation was significantly reduced when FMRP was not pre-incubated with the translation template (Fig. 3B). This raises an intriguing possibility that in living cells some mRNA species may be pre-disposed to translation suppression by FMRP, either due to an intrinsic high affinity for FMRP or to pre-exposure to FMRP prior to other RNPs, perhaps during transcription in the nucleus. This may provide a mechanism for FMRP to preferentially influence translation of some mRNA species as compared to others in living cells.
It is important to note that RRL is a simplified system, although it has been extensively used to study translation regulation by a variety of RNA-binding proteins (18,22,29). It is likely that the influence of FMRP on protein synthesis in vivo may be modulated by sophisticated mechanisms that do not exist in RRL. However, as a basic approach to functional inquiry the RRL system does provide novel insights into the function of FMRP. Identification of the physiological mRNA targets for FMRP should greatly facilitate understanding of the function of FMRP, as well as the pathogenesis of fragile X syndrome. In this regard, the in vitro translation assay described above can be readily used to directly test the influence of FMRP on translation of its physiological target mRNAs and possible mechanisms involved once these mRNAs are identified.
ACKNOWLEDGEMENTS
We thank Drs G.Dreyfuss and J.-L.Mandel for generously providing antibodies and Dr Silver and Dr Campagnoni for cDNA constructs. This work was supported in part by NIH grant HD35576 to Y.F. and K.D.W. and by NIH grant HD20521 to S.T.W. S.T.W. is an Investigator of the Howard Hughes Medical Institute.
To whom correspondence should be addressed. Tel: +1 404 727 0351; Fax: +1 404 727 0365; Email: yfeng@emory.edu
Figure 1. Dose-dependent reduction of translation of brain poly(A) RNA but not rabbit reticulocyte poly(A) RNA by FMRP. (A) Dose-dependent effect of FMRP. An aliquot of 1.5 µg poly(A) RNA was used in each reaction. Translation yield for each reaction is represented by the percentage of TCA precipitable counts (c.p.m.) obtained from mock-treated reactions. At 10 min after initiation of translation yields for reactions exposed to various amounts of FMRP or BSA as depicted at the bottom were subjected to one way ANOVA analysis (P < 0.001). The number of experiments for each treatment is also depicted at the bottom. The asterisks indicate P values (***, P < 0.001) in comparison to the yield from the mock-treated reaction. (B) SDS–PAGE analysis of the translation inhibition caused by a high concentration of FMRP. FMRP concentration in the reaction was 250 nM, as depicted at the top of the corresponding lane. (C) Coomassie Blue staining of a SDS–PAGE gel of FMRP and BSA used in the translation reaction. The amount of protein loaded is indicated at the top of the corresponding lane. (D) Recombinant FMRP remains intact during translation. Translation mix containing various amounts of Flag–FMRP, as indicated at the top of the corresponding lanes, was subjected to SDS–PAGE immunoblotting at the end of the reaction by anti-Flag antibody.
Figure 2. FMRP does not accelerate degradation of mRNA templates. The concentration of FMRP in translation reactions was 250 nM. Signals for corresponding RNAs are indicated on the left and the presence/absence of FMRP is indicated at the top. GAPDH, glyceraldehyde phosphate dehydrogenase; RPS14, ribosomal protein S14; MBP, myelin basic protein. (A) Northern hybridization of FMRP-treated and mock-treated RNA templates before translation in RRL. (B) Northern hybridization of FMRP-treated and mock-treated brain poly(A) RNA recovered from RRL after translation.
Figure 3. Interaction of FMRP with mRNPs in RRL may modulate the potency of FMRP as a translation inhibitor. (A) Co-immunoprecipitation of FMRP with FXR2P from RRL. Translation reactions were carried out with 250 nM FMRP when indicated (+). Immunoprecipitations with and without anti-M2 beads were carried out as described in Materials and Methods. An aliquot of flow-through, last wash and eluate for each reaction was loaded on a SDS–PAGE gel followed by immunoblot analysis, using anti-FXR2 antibody. (B) Translation inhibition of brain poly(A) RNA with and without pre-exposure to FMRP. Translation yield of each reaction at 10 and 20 min was plotted. The amount of and treatment by FMRP as well as the number of experiments for each treatment are indicated at the bottom. ***, P < 0.001 for translation yield compared to mock-treated reaction; +++, P < 0.001 for translation yield comparing FMRP treatment with and without pre-incubation with mRNA.
Figure 4. Translation inhibition by FMRP can be reversed by the Fmr1 3′-UTR. An aliquot of 90 nM rat cerebral cortical poly(A) RNA was used in each reaction. Ten nanomolar Fmr1 3′-UTR or the antisense transcript of β-globin cDNA was included when indicated. An aliquot of 125 nM FMRP was used in the reactions when indicated. The treatment of brain mRNA templates is indicated in the upper left panel. n, experiment number; **, P < 0.01 in comparison to translation yield derived from the mock-treated reaction.
Figure 5. FMRP inhibits translation of PTH but not globin message. (A) Dose-dependent translation inhibition by FMRP on PTH RNA but not on rabbit reticolucyte poly(A) RNA. 35S-labeled translation products were fractionated on a SDS–PAGE gel before being subjected to phosphorimager analysis. The concentration of FMRP (nM) used in each reaction is indicated at the top of the corresponding lanes, with the 35S-labeled peptide indicated on the left. The translation yield of PTH on SDS–PAGE was quantitatively analyzed with a phosphorimager. Results from two separate experiments were averaged and are indicated at the bottom of the corresponding lanes. (B) FMRP selectively inhibits translation of PTH mRNA but not rabbit reticulocyte poly(A) RNA when present in the same reaction. The presence/absence of FMRP is indicated at the top of the corresponding lanes. The concentration of FMRP was 250 nM in the indicated reaction. (C) FMRP treatment does not cause degradation of PTH transcripts. Total RNA was recovered from FMRP-treated or mock-treated translation reactions at various time points of translation as indicated at the top of each lane. Northern hybridization was performed using 32P-labeled probes as indicated. 28S rRNA was used as a loading control. (D) FMRP selectively binds PTH transcript but not β-globin transcript. An in vitro RNA binding assay was carried out as described in Materials and Methods. The biotinylated RNA used in each reaction was 20 nM based on the OD260 reading and confirmed by ethidium bromide stained agarose gel electrophoresis.
Figure 6. Removing the FMRP-binding site on the mRNA abolishes the effect of FMRP on translation. (A) FMRP binds to the MBP 3′-UTR. A RNA binding assay was carried out as described in Materials and Methods. The bar graph presents results based on scintillation counting of bound [35S]FMRP and asterisks indicate P values in paired t-tests (***, P < 0.001; n = 4). The insert illustrates a phosphorimage of [35S]FMRP captured by the full-length and the 3′-UTR-truncated MBP transcripts fractionated by SDS–PAGE. (B) Removing the MBP 3′-UTR abolished the inhibitory effect of FMRP on MBP translation. An aliquot of 125 nM recombinant FMRP and an equimolar concentration of capped MBP transcript were used in the reaction when indicated. The translation yield was determined by scintillation counting of TCA precipitable c.p.m. as described in the legend to Figure 1. Asterisks indicate P values in paired t-tests (***, P < 0.001; n = 4) in comparison to the yield from the mock-treated reaction.
References
1 Warren,S.T. and Nelson,D.L. (
2 Eberhart,D.E. and Warren,S.T. (
3 de Vries,B.B., Halley,D.J., Oostra,B.A. and Niermeijer,M.F. (
4 Jin,P. and Warren,S.T. (
5 Ashley,C.T., Wilkinson,K.D., Reines,D. and Warren,S.T. (
6 Siomi,H., Siomi,M.C., Nussbaum,R.L. and Dreyfuss,G. (
7 Brown,V., Small,K., Lakkis,L., Feng,Y., Gunter,C., Wilkinson,K.D. and Warren,S.T. (
8 Corbin,F., Bouillon,M., Fortin,A., Morin,S., Rousseau,F. and Khandjian,E.W. (
9 Feng,Y., Absher,D., Eberhart,D.E., Brown,V., Malter,H.E. and Warren,S.T. (
10 Tamanini,F., Meijer,N., Verheij,C., Willems,P.J., Galjaard,H., Oostra,B.A. and Hoogeveen,A.T. (
11 Feng,Y., Gutekunst,C.A., Eberhart,D.E., Yi,H., Warren,S.T. and Hersch,S.M. (
12 Zhang,Y., O’Connor,J.P., Siomi,M.C., Srinivasan,S., Dutra,A., Nussbaum,R.L. and Dreyfuss,G. (
13 Ceman,S., Brown,V. and Warren,S.T. (
14 Bardoni,B., Schenck,A. and Mandel,J.L. (
15 Eberhart,D.E., Malter,H.E., Feng,Y. and Warren,S.T. (
16 Sittler,A., Devys,D., Weber,C. and Mandel,J.-L. (
17 Siomi,M.C., Zhang,Y., Siomi,H. and Dreyfuss,G. (
18 Ostareck,D.H., Ostareck-Lederer,A., Wilm,M., Thiele,B.J., Mann,M. and Hentze,M.W. (
19 Jan,E., Motzny,C.K., Graves,L.E. and Goodwin,E.B. (
20 Saccomanno,L., Loushin,C., Jan,E., Punkay,E., Artzt,K. and Goodwin,E.B. (
21 Craig,A.W., Haghighat,A., Yu,A.T. and Sonenberg,N. (
22 Evdokimova,V.M., Kovrigina,E.A., Nashchekin,D.V., Davydova,E.K., Hershey,J.W. and Ovchinnikov,L.P. (
23 Ashley,C.T., Sutcliffe,J.S., Kunst,C.B., Leiner,H.A., Eichler,E.E., Nelson,D.L. and Warren,S.T. (
24 Kafatos,F.C., Efstratiadis,A., Forget,B.G. and Weissman,S.M. (
25 Moallem,E., Kilav,R., Silver,J. and Naveh-Many,T. (
26 Campagnoni,A.T., Hunkeler,M.J. and Moskaitis,J.E. (
27 Feng,Y., Lakkis,L., Devys,D. and Warren,S.T. (
28 Gonzalez,K.H., Blomquist,H. and Holmgren,G. (
29 Laggerbauer,B., Ostareck,D., Keidel,E.M., Ostareck-Lederer,A. and Fischer,U. (
30 Adinofi,S., Bagni,C., Musco,G., Gibson,T., Mazzarella,L. and Pastore,A. (
31 Tamanini,F., Willemsen,R., van Unen,L., Bontekoe,C., Galjaard,H., Oostra,B.A. and Hoogeveen,A.T. (
32 Hinds,H.L., Ashley,C.T., Nelson,D.L., Warren,S.T., Housman,D.E. and Schalling,M. (
33 Devys,D., Lutz,Y., Rouyer,N., Bellocq,J.-P. and Mandel,J.-L. (
34 Khandjian,E.W., Fortin,A., Thibodeau,A., Tremblay,S., Cote,F., Devys,D., Mandel,J.-L. and Rousseau,F. (
35 Peier,A.M., McIlwain,K.L., Kenneson,A., Warren,S.T., Paylor,R. and Nelson,D.L. (



![Figure 6. Removing the FMRP-binding site on the mRNA abolishes the effect of FMRP on translation. (A) FMRP binds to the MBP 3′-UTR. A RNA binding assay was carried out as described in Materials and Methods. The bar graph presents results based on scintillation counting of bound [35S]FMRP and asterisks indicate P values in paired t-tests (***, P < 0.001; n = 4). The insert illustrates a phosphorimage of [35S]FMRP captured by the full-length and the 3′-UTR-truncated MBP transcripts fractionated by SDS–PAGE. (B) Removing the MBP 3′-UTR abolished the inhibitory effect of FMRP on MBP translation. An aliquot of 125 nM recombinant FMRP and an equimolar concentration of capped MBP transcript were used in the reaction when indicated. The translation yield was determined by scintillation counting of TCA precipitable c.p.m. as described in the legend to Figure 1. Asterisks indicate P values in paired t-tests (***, P < 0.001; n = 4) in comparison to the yield from the mock-treated reaction.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/nar/29/11/10.1093/nar/29.11.2276/2/m_gke33906.jpeg?Expires=1716438612&Signature=o0l8V1sMlzQap0VU2HgvFp9p8-g5bSdIu9ECYMpUkPIsFRS2458pjKqEsnvIE7zNdU0fGAjA4M6H694J1TvKQh61KXa5NGsSsEGOUkqh8~6~h9aW-vLvDTUPbKs6dzDKrWzIJk1~q7X9f~kJietzsTFIYsMUItKq-9LVeUqBd3zB1D61-XehjmhpStSyPbKInQooX28eRNpMctT9J1E0R~-iX37t2YqXBTjkliV8ZcNNjeL1y2ev2KFgKvRvBNb-cZbGIxUKlWAHR~Vw8~7H5Wyde0O84C1YaImz6jBHoCkTmJeViAmuaGZSSkpS62Rk1eUe-I1XpbbdxlftIa5YAw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

Comments