Abstract
In addition to apolipoprotein E (APOE), recent large genome-wide association studies (GWASs) have identified nine other genes/loci (CR1, BIN1, CLU, PICALM, MS4A4/MS4A6E, CD2AP, CD33, EPHA1 and ABCA7) for late-onset Alzheimer's disease (LOAD). However, the genetic effect attributable to known loci is about 50%, indicating that additional risk genes for LOAD remain to be identified. In this study, we have used a new GWAS data set from the University of Pittsburgh (1291 cases and 938 controls) to examine in detail the recently implicated nine new regions with Alzheimer's disease (AD) risk, and also performed a meta-analysis utilizing the top 1% GWAS single-nucleotide polymorphisms (SNPs) with P<0.01 along with four independent data sets (2727 cases and 3336 controls) for these SNPs in an effort to identify new AD loci. The new GWAS data were generated on the Illumina Omni1-Quad chip and imputed at ∼2.5 million markers. As expected, several markers in the APOE regions showed genome-wide significant associations in the Pittsburg sample. While we observed nominal significant associations (P<0.05) either within or adjacent to five genes (PICALM, BIN1, ABCA7, MS4A4/MS4A6E and EPHA1), significant signals were observed 69–180 kb outside of the remaining four genes (CD33, CLU, CD2AP and CR1). Meta-analysis on the top 1% SNPs revealed a suggestive novel association in the PPP1R3B gene (top SNP rs3848140 with P=3.05E–07). The association of this SNP with AD risk was consistent in all five samples with a meta-analysis odds ratio of 2.43. This is a potential candidate gene for AD as this is expressed in the brain and is involved in lipid metabolism. These findings need to be confirmed in additional samples.
Similar content being viewed by others
Introduction
Alzheimer's disease (AD), especially the late-onset form (LOAD), is a complex multifactorial neurodegenerative disease and the leading cause of dementia among the elderly people.1 Currently, there are ∼5 million AD cases in the United States and about 35 million cases worldwide.2 Due to its long clinical course, AD is a major public health problem. Genetic susceptibility at multiple genes and interactions among them and/or environmental factors likely influence the risk of AD. AD has a strong genetic basis with heritability estimates up to 80%.3 However, genetic variants in the four well-established genes for AD; amyloid precursor protein and presenilin 1 and 2 (PSEN1 and PSEN2) for the rare early-onset, and apolipoprotein E (APOE) for the common LOAD explain less than half of this heritability.
To identify the remaining genes for the common LOAD, efforts have been focused on conducting genome-wide association studies (GWASs)4, 5, 6, 7, 8, 9, 10, 11 because this approach is hypothesis free and conceptually would identify all known and unknown genes. However, with the exception of the APOE region, no other significant associations were replicated across these initial GWAS. This highlights the difficulties in identifying the remaining LOAD genes that are thought to make a relatively small contribution to the overall risk of disease and thus would require much large sample size than used in earlier GWASs.4, 5, 6, 7, 8, 9, 10, 11 Recent large GWASs have identified nine additional genes/loci for LOAD, including CR1, BIN1, CLU (a.k.a. APOJ), PICALM, MS4A4/MS4A6E, CD2AP, CD33, EPHA1 and ABCA7.12, 13, 14, 15, 16 Although up to 80% of the AD risk is attributable to genetic factors,3 all of the known LOAD genes (including APOE and new ones) account for about 50% of the total genetic variance. This indicates that additional risk genes for LOAD remain to be identified.
In this study, we used 2229 samples from the University of Pittsburgh Alzheimer's Disease Research Center (ADRC) that were genotyped using the Illumina Omni1-Quad chip to examine the extent of associations of the recently implicated nine non-APOE risk loci for LOAD in this sample. In addition, we performed a GWAS analysis in the Pittsburgh sample and conducted a meta-analysis on the top 1% single-nucleotide polymorphisms (SNPs) by incorporating additional 2727 cases and 3336 controls from prior studies to identify new loci for AD.
Subjects and methods
Stage 1 sample
Genomic DNA from 1440 AD cases and 1000 controls were genotyped using the Illumina Human Omni1-Quad as part of the stage 1 discovery sample. All subjects were European Americans. AD cases (mean age-at-onset (AAO) 72.6±6.4 years; 65.6% women; 23.5% autopsy-confirmed) were derived from the University of Pittsburgh ADRC, all of whom met the National Institute of Neurological and Communication Disorders and Stroke (NINCDS)/Alzheimer's Disease and Related Disorders Association (ADRDA) criteria for probable or definite AD. The University of Pittsburgh ADRC follows a standard evaluation protocol, including medical history, general medical and neurological examinations, psychiatric interview, neuropsychological testing and magnetic resonance imaging scan. Controls were cognitively normal subjects that were recruited from the same geographical area as the cases. They were 60 years or older (mean age 74.07±6.20 years; 59.8% women; 0.2% autopsy-confirmed), had no psychiatric or neurological disorders, and did not meet criteria for mild cognitive impairment or dementia. All subjects were recruited with informed consent, and the study was approved by the University of Pittsburgh Institutional Review Board.
Stage 2 samples
A total of 2727 cases and 3336 controls were derived from four prior studies and they are briefly described below.
Mayo sample
The Mayo sample comprised 844 AD cases (mean AAO: 74±4.8 years; 57.2% women) and 1255 controls (mean age: 73.2±4.4 years; 51.7% women) that were previously genotyped using the Illumina HumanHap300 BeadChip.11 AD diagnosis was established using the NINCDS/ADRDA criteria.
Alzheimer's Disease Neuroimaging Initiative (ADNI) sample
This data set consisted of 188 AD cases with a clinical diagnosis of AD at baseline visit and 193 controls (mean age: 78.6±5.3 years; 46.3% women) that were previously genotyped using the Illumina 610-Quad BeadChip.17 AD cases were between the ages of 55 and 90 (mean AAO: 71.9±8.1 years; 44.6% women) and met NINCDS/ADRDA criteria for AD. Details of the clinical evaluation and sample characterization are described elsewhere.18, 19 The ADNI data used in this report were obtained from the ADNI database (adni.loni.ucla.edu). The initial goal of ADNI was to recruit 800 adults, aged 55–90, to participate in research on the sensitivity and specificity of neuroimaging and other biomarkers for detecting and monitoring AD pathology in vivo. In ADNI, ∼200 cognitively normal older individuals and 400 people with amnestic mild cognitive impairment were followed for 3 years and 200 people with mild early-stage AD who were followed for 2 years. For up-to-date information, see http://www.adni-info.org.
University of Miami/Vanderbilt University/Mt. Sinai School of Medicine sample
The University of Miami/Vanderbilt University/Mt. Sinai School of Medicine data set contains 1186 cases (mean AAO: 74.1±7.8 years; 64% women) and 1135 controls (mean age: 74.0±8.3; 61% women) ascertained at the University of Miami, Vanderbilt University and Mt. Sinai School of Medicine. Cases met NINCDS/ADRDA criteria for probable or definite AD with AAO >60 years. Cognitively healthy controls were unrelated individuals from the same catchment areas.16
MIRAGE study sample
MIRAGE is a family-based genetic epidemiological study of AD that enrolled AD cases and unaffected sibling controls at 17 clinical centers in the United States, Canada, Germany and Greece. Briefly, families were ascertained through a proband meeting the NINCDS/ADRDA criteria for definite or probable AD. Unaffected sibling controls were verified as cognitively healthy based on a Modified Telephone Interview of Cognitive Status score of ⩾86. For this study, we used 1262 subjects (509 cases and 753 controls)16 from the MIRAGE Caucasian data set.
Genotyping and quality control of genotype data in the Pittsburgh sample
The stage 1 Pittsburgh sample was genotyped using the Illumina Omni1-Quad chip (containing probes for 1 016 423 SNPs and/or copy-number variations) at the Feinstein Institute of Medical Research (Manhasset, NY). Genotypes for two APOE SNPs rs429358 (E*4) and rs7412 (E*2) were determined either as previously described20 or using TaqMan SNP genotyping assays.
Exclusion criteria for individual samples included consent and diagnostic criteria (five controls were excluded because of updated diagnostic criteria, or refusal to participate further in this study), high genotype failure rate (141 individuals were removed because of a genotype failure rate >2%) and cryptic relatedness (65 individuals were removed because they displayed an average degree of sharing (identity by state or IBS) >0.4 with other members of the data set). Exclusion criteria for markers included minor allele frequency (189 727 SNPs were removed because of minor allele frequency <1%), deviation from Hardy–Weinberg expectations in controls (2239 SNPs gave a Hardy–Weinberg expectations test P-value ⩽1E–06), and high genotype failure rates (22 385 SNPs were removed because of genotype failure rates >2%). The final sample after all exclusions was applied consisted of 1291 cases and 938 controls.
Imputation
Genotype posterior probabilities were imputed with MACH v.1.0 (http://www.sph.umich.edu/csg/abecasis/MACH/), on all data sets using haplotypes from the HapMap CEU v3 data release as a reference sample. The imputation generated data for >3 million SNPs that were subsequently filtered to exclude SNPs with r2<0.3 and 2 543 888 were included in the analysis. Additional details of the imputation procedure were published previously.16
Population stratification
Population stratification was examined using a multi-dimensional scaling method as implemented in PLINK.21 Four components were conservatively determined to be relevant to the determination of population origin based on the visual examination of principle component plots.
Association analyses
Initial association analysis was performed in the University of Pittsburgh ADRC's GWAS data set. After standard quality control filters for both genotypes and samples and imputing for unobserved genotypes, a total of ∼2.5 million SNPs were examined in 2229 subjects (1291 cases and 938 controls) for association analysis. Association of SNPs with AD risk was tested using logistic regression under an additive model that included age, sex and the first four principal components as covariates. All SNPs with P-values of <0.01 were carried forward to the meta-analysis. Significance values from logistic regression analyses were used for ranking purposes only, and so were not adjusted for multiple testing. All analyses were done in R and/or PLINK. Meta-analysis was done using a fixed-effect methodology, as implemented in PLINK. Heterogeneity testing was accomplished using Cochran's Q statistic, summarized as the I2 statistic (the percentage of total variation across studies that is due to heterogeneity rather than chance).22, 23 Values closer to 0 indicate no heterogeneity, whereas larger numbers indicate increasing degrees of heterogeneity between studies.
Results
Association analysis in nine non-APOE AD loci
We first examined the association signals in the Pittsburgh GWAS data in recently reported nine non-APOE AD gene regions (CR1, BIN1, CLU, PICALM, MS4A4/MS4A6E, CD2AP, CD33, EPHA1 and ABCA7)12, 13, 14, 15, 16 in 2229 cases and controls that passed quality control for both genotypes and samples and included imputed data for unobserved genotypes. Although this sample was included as one of several replication samples in one GWAS that reported the latter five gene region,16 the extent of replications in these five and the other four regions in this sample has previously not been examined closely. We observed significant associations (P<0.05) either within or adjacent to five genes (PICALM, BIN1, ABCA7, MS4A4/MS4A6E and EPHA1). On the other hand, significant signals were observed 69–180 kb outside of the remaining four genes (CD33, CLU, CD2AP and CR1). The regional association plots containing SNPs within ∼500 kb on either side of these nine genes are presented in Supplementary Figures 1–9.
In the PICALM gene and its surrounding sequence, 69 SNPs showed significant associations (Supplementary Figure 1). The top 11 SNPs with P<E–03 were located in four different introns of the PICALM gene. The most significant SNP, rs17817992 (P=1.37E–04), was located in intron 2 followed by rs12790526 (P=1.43E–04) in intron 2, rs12795381 (P=1.68E–04) in intron 12 and rs12802399 (P=1.72E–04) in intron 2. The originally reported genome-wide significant SNP/rs3851179,13 which is present about 89 kb 5′ to PICALM, was observed at P=4.6E–02 in our sample. In the BIN1 gene, 17 SNPs showed significant associations (Supplementary Figure 2), mostly in the 5′region, including the 5 top significant SNPs/rs11680911 (P=2.9E–04), rs7561528 (P=3.0E–04), rs6743470 (P=1.18E–03), rs17014923 (P=1.64E–03) and rs9394826 (P=2.15E–03). Among our top five significant SNPs, rs7561528 was genome-wide significant in the discovery sample of the Alzheimer Disease Genetics Consortium.16 The originally reported genome-wide significant SNP/rs744373 in the initial GWAS after stage 3 meta-analysis14 was observed at P=1.02E–02 in our sample. In the ABCA7 gene, seven SNPs showed significant association (Supplementary Figure 3), including the two most significant SNPs, rs4147916 in intron 34 (P=3.18E–03) and rs4147918 in exon 37 (P=3.19E–03) that is associated with an amino-acid change (Gln → Arg). The reported genome-wide significant SNP (rs3764650)15 gave a non-significant but similar trend of association in our sample (P=1.68E–01).
There are multiple genes in the MS4A gene clusters on chromosome 11. While one GWAS reported genome-wide significant SNPs in the MS4A4A gene,16 another GWAS found genome-wide significant SNPs in the MS4A6A and MS4A4E genes.15 In our data set, we found a total of 39 SNPs with P<0.05 in the MS4A gene cluster (Supplementary Figure 4) and the 5 most significant SNPs were located in the MS4A4A gene and its surrounding sequence: rs11827324 (P=2.58E–03), rs4939338 (P=3.39E–03), rs10792263 (P=4.85E–03), rs11824773 (P=8.06E–03) and rs11824734 (P=6.93E–03). The genome-wide significant SNP in the Alzheimer Disease Genetics Consortium (rs4938933) gave the same trend of association in our sample (P=8.80E–02). There were eight significant SNPs in the EPHA1 gene (Supplementary Figure 5), including the top significant SNPs: rs10241042 (P=2.27E–03), rs1525119 (P=1.02E–02) and rs10233030 (P=1.29E–02). The reported genome-wide significant SNP (rs11767557)15, 16 was observed with a similar trend of association in our sample (P=1.11E–01).
For the other four genes, significant SNPs (P<0.05) were observed 69, 125, 143 and 180 kb outside of the CD33, CLU, CR1 and CD2AP genes, respectively. There were four SNPs with P<0.05 and another seven with P-values between 0.05 and 0.10 located in an intergenic region about 69 kb distal to the CD33 gene. Likewise, there were four significant SNPs (P=0.012–0.027) about 125 kb proximal to the CD33 gene (Supplementary Figure 6). The reported genome-wide significant SNP (rs3865444)15, 16 gave a non-significant but similar trend of association in our sample (P=2.50E–01). Overall, recombination was found to be low in this region. In the CLU region, 41 significant SNPs were present in the PTK2B and CHRNA2 genes, both of which are within 100–125 kb of the CLU gene (Supplementary Figure 7). Although no SNP showed an association at P=0.05 in the CLU gene, several SNPs demonstrated the reported trend of association. The 14 most significant SNPs were present in the PTK2B gene with P<4.0E–03. The top three SNPs in the PTK2B gene were rs373625 in intron 25 (P=2.72E–03) and rs9657295 and rs11135993 in intron 27 (P=3.06E–03). The reported associations in the CD2AP and CR1 genes were not replicated in our sample, as significant SNPs were located ∼150 kb from these two genes (Supplementary Figures 8 and 9).
GWAS analysis
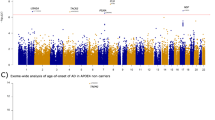
We next examined the entire GWAS data set (∼2.5 million SNPs) in the Pittsburg sample. Figure 1a shows the comparison of observed P-values with the P-values for a null distribution. The observed P-value conformed to the null distribution until the tail of the distribution where it deviated, suggesting little evidence of stratification but compelling evidence of disease associations. As expected, several SNPs in the APOE region on chromosome 19 demonstrated genome-wide significant (P<5E–08) associations (Figure 2). The most significant SNP was rs429358 (E*4) in the APOE gene (P=2.52E–53) followed by rs4420638 (P=1.97E–42), rs6857 (P=6.75E–38), rs2075650 (P=5.67E–29), rs157582 (P=5.56E–28) and rs157580 (P=2.77E–10) in the APOE/APOC1/TOMM40 gene region. The regional association plot including all SNPs in the APOE region is given in Figure 3.
Quantile-quantile plots showing the observed versus the expected P-values in the University of Pittsburgh sample including all single-nucleotide polymorphisms (SNPs) (a) and after removing SNPs in the APOE, CR1, BIN1, CLU, PICALM, MS4A4/MS4A6E, CD2AP, CD33, EPHA1 and ABCA7 regions (b). The red line shows the distribution under the null hypothesis.
Manhattan plots showing the genome-wide P-values in the University of Pittsburgh sample after adjusting for age (age-at-onset in cases), sex and principle components. Red line indicates genome-wide significant level (P<5E–08) and blue line indicates suggestive associations (P<1E–04) in the Pittsburgh sample.
After removing SNPs in the APOE, CR1, BIN1, CLU, PICALM, MS4A4/MS4A6E, CD2AP, CD33, EPHA1 and ABCA7 regions, a deviation of P-values from the null distribution remained in the quantile-quantile plot, although within the 95% confidence interval of the expectation (Figure 1b). Since no other SNPs outside the APOE region were genome-wide significant in the Pittsburgh discovery sample, we performed a meta-analysis by combining the results of the top ∼1% SNPs (∼25 000) with P<0.01 of the discovery sample with the four existing data sets (2727 AD cases and 3336 controls) for these SNPs. Of the known loci, APOE, BIN1 and PICALM revealed genome-wide significant associations along with five suggestive new loci with P<1E–05 in the meta-analysis (Table 1). All SNPs with P<1E–04 in the meta-analysis are presented in Supplementary Table.
Four of the top five SNPs outside the APOE and nine other known loci were either directly genotyped or had proxy genotyped SNPs with P<1E–04 on Illumina arrays (see Table 1 and Supplementary Table), thus eliminating potential spurious associations due to imputation artifact. The most significant SNP, rs3848140 (P=3.05E–07), located in the PPP1R3B (encoding protein phosphatase 1, regulatory subunit 3B) region on chromosome 8 at position 9.04 Mb was actually genotyped and there were three additional significant SNPs in this gene at P<1E–06. The association of this SNP with AD risk was consistent in all five samples with meta odds ratio of 2.43. Figure 4 shows the regional plot for SNPs within 500 kb on either side of the PPP1R3B index SNP and the meta P-values for markers which had P<IE–04 in the meta-analysis. The regional plots for the remaining four top AD loci are shown in Supplementary Figures 10–13. To remove any variation in AD due to the established effect of APOE, we also analyzed the data after adjusting for the effect of APOE*4, but found no appreciable difference in P-values for the non-APOE loci (Table 1), indicating that their effects are independent of APOE.
Discussion
In this study, we have used a new GWAS data set to (i) examine in detail the recently implicated nine new regions with AD risk, and to (ii) perform a meta-analysis utilizing the top 1% GWAS SNPs (∼25 000) with P<0.01 along with four independent data sets for these SNPs in an effort to identify new AD loci.
Five of the nine loci are replicated in the Pittsburgh sample as we found multiple nominal significant SNPs in or around five genes (PICALM, BIN1, ABCA7, MS4A4/MS4A6E and EPHA1), suggesting that these are the relevant genes for AD in these regions. As associations in these genes have been validated in multiple studies,13, 16 it is acceptable to consider P<0.05 statistically significant in follow-up studies24 like ours. However, the identity of the potential causal genes in the remaining four regions (CD33, CLU, CD2AP and CR1) is not clear in the Pittsburgh sample, as significant SNPs were observed 69–180 kb outside of these genes. The power in our Pittsburgh sample to detect these nine loci for the effect sizes reported in a previous GWAS16 is 0.38, 0.43, 0.39, 0.31, 0.38, 0.30, 0.28, 0.28 and 0.47 for PICALM, BIN1, ABCA7, MS4A4, EPHA1, CD33, CLU, CD2AP and CR1, respectively. Although the overall power is low and this would reduce our confidence in our negative results, it enhances the interest in our positive results even further.
There were only 11 SNPs present in the CD33 gene on the 1M chip. This may explain why we did not observe any significant SNP in this gene, although the reported genome-wide significant SNP (rs3865444) gave a P-value of 0.25 with the same effect size in our sample. Also, recombination is low in the CD33 region; thus, it is likely that functional SNPs may be present in a broader region around CD33. Although none of the small number of 15 SNPs present in the CLU gene on the 1M chip showed significant association at P<0.05, a number of them showed the trend of association reported in earlier studies. However, several SNPs in a nearby PTK2B gene that is expressed in adult brain,25 revealed significant associations. Perhaps, genetic variations in the CLU and PTK2B genes, both of which are expressed in the brain, are relevant to AD risk and thus resequencing a broader region in and around these genes may reveal functional variants. However, a recent sequencing study found no functional SNPs in the coding region of the CLU gene and, likewise, no significant association was found between the previously associated CLU SNPs with AD risk with expression quantitative trait loci,26 suggesting that the biological role of CLU is AD is not yet clear. While findings in our sample suggest the involvement of broader regions around the CD33 and CLU genes, the evidence of association of the CR1 and CD2AP with AD is weak or absent.
The GWAS analysis in the Pittsburgh data set found only the APOE/TOMM40/APOC1 region to be genome-wide significant, which is due to the established association with the APOE*4 SNP. Meta-analysis of the top 25 000 SNPs from the Pittsburgh GWAS data, in conjunction with four replication data sets revealed five suggestive non-APOE loci. The top SNP was located in the PPP1R3B gene, which is a potential candidate gene for AD as it is expressed in human brain.27 Genetic variation in PPP1R3B has been shown to be associated with plasma high density lipoprotein and cholesterol levels and the risk of coronary artery disease.28, 29 Overexpression of the mouse ortholog Ppp1r3b in mouse liver results in significantly lower high density lipoprotein levels.28 As high density lipoproteins levels and particle size affect the clearance of β-amyloid in mouse brain,30 it is conceivable that the PPP1R3B-associated effect on AD risk is mediated through its impact on high density lipoproteins levels. Although the association of the top PPP1R3B SNP did not meet the strict criteria for genome-wide significance, it was consistent in direction in all samples as reflected in Odds ratio values (see Table 1). This suggests that this gene region should be followed up in additional samples. If these findings are confirmed, then this would be another cholesterol-related gene involved in AD along with APOE, CLU and ABCA7.
In conclusion, 6 of the 10 established AD loci, including the APOE locus, have been replicated in our new GWAS data set, while broader regions in the remaining four loci are suspected. Although we did not identify additional non-APOE SNPs meeting a conservative threshold of genome-wide significance, we have identified five suggestive loci that warrant follow-up in additional samples.
References
Evans DA, Funkenstein HH, Albert MS, Scherr PA, Cook NR, Chown MJ et al. Prevalence of Alzheimer′s disease in a community population of older persons. Higher than previously reported. JAMA 1989; 262: 2551–2556.
Alzheimer's Disease International. World Alzheimer Report 2009. www.alz.co.uk/research/world-report.
Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 2006; 63: 168–174.
Grupe A, Abraham R, Li Y, Rowland C, Hollingworth P, Morgan A et al. Evidence for novel susceptibility genes for late-onset Alzheimer′s disease from a genome-wide association study of putative functional variants. Hum Mol Genet 2007; 16: 865–873.
Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Lince DH et al. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer′s disease. J Clin Psychiatry 2007; 68: 613–618.
Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, Zismann VL et al. GAB2 alleles modify Alzheimer′s risk in APOE epsilon4 carriers. Neuron 2007; 54: 713–720.
Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol 2008; 65: 45–53.
Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF et al. Genome-wide association analysis reveals putative Alzheimer′s disease susceptibility loci in addition to APOE. Am J Hum Genet 2008; 83: 623–632.
Beecham GW, Martin ER, Li YJ, Slifer MA, Gilbert JR, Haines JL et al. Genome-wide association study implicates a chromosome 12 risk locus for late-onset Alzheimer disease. Am J Hum Genet 2009; 84: 35–43.
Abraham R, Moskvina V, Sims R, Hollingworth P, Morgan A, Georgieva L et al. A genome-wide association study for late-onset Alzheimer′s disease using DNA pooling. BMC Med Genomics 2008; 1: 44.
Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP et al. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer's disease. Nat Genet 2009; 41: 192–198.
Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer′s disease. Nat Genet 2009; 41: 1094–1099.
Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer′s disease. Nat Genet 2009; 41: 1088–1093.
Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 2010; 303: 1832–1840.
Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer′s disease. Nat Genet 2011; 43: 429–435.
Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer′s disease. Nat Genet 2011; 43: 436–441.
Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S et al. Alzheimer′s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: genetics core aims, progress, and plans. Alzheimers Dement 2010; 6: 265–273.
Aisen PS, Petersen RC, Donohue MC, Gamst A, Raman R, Thomas RG et al. Clinical core of the Alzheimer′s Disease Neuroimaging Initiative: progress and plans. Alzheimers Dement 2010; 6: 239–246.
Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ et al. Alzheimer′s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology 2010; 74: 201–209.
Kamboh MI, Aston CE, Hamman RF . The relationship of APOE polymorphism and cholesterol levels in normoglycemic and diabetic subjects in a biethnic population from the San Luis Valley, Colorado. Atherosclerosis 1995; 112: 145–159.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet 2007; 81: 559–575.
Brockwell SE, Gordon IR . A comparison of statistical methods for meta-analysis. Stat Med 2001; 20: 825–840.
Higgins JC, Ivanov NV, Hodor P, Xia M, Wei N, Blevins R . et al. Measuring inconsistency in meta-analyses. BMJ 2003; 327: 557–560.
Plenge RM, Bridges SL, Huizinga WJ, Criswell LA, Gregersen PK . Recommendation of genetic association studies in Arthritis & Rheumatism. Arthritis Rheum 2001; 63: 2839–2847.
Lev S, Moreno H, Martinez R, Canoll P, Peles P, Musacchio JM et al. Proteintyrosine kinase PYK2 involved in Ca(2+)-induced regulation of ion channel and MAP kinase functions. Nature 1995; 376: 737–745.
Guerreiro RJ, Beck J, Gibbs JR, Santana I, Rossor MN, Schott JM et al. Genetic variability in CLU and its association with Alzheimer's diseases. PLoS One 2010; 5: e9510.
Glorioso C, Oh S, Douillard GG, Sibille E . Brain molecular aging, promotion of neurological disease and modulation by sirtuin 5 longevity gene polymorphism. Neurobiol Dis 2011; 41: 279–290.
Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010; 466: 707–713.
Waterworth DM, Ricketts SL, Song K, Chen L, Zhao JH, Ripatto S et al. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol 2010; 30: 2264–2276.
Cramer PE, Cirrito JR, Wesson DW, Lee CYD, Karlo C, Zinn AE et al. ApoE-directed therapeutics rapidly clear β-amyloid and reverse deficits in AD mouse models. Science 2012; 335: 1503–1506.
Acknowledgements
This study was supported by the National Institute on Aging grants AG030653 (MIK), AG005133 (ADRC), AG027224 (RAS), AG07562 and AG023651 (MG) and AG18023 (SGY). We thank Drs Peter Gregersen and Annette Lee of the Feinstein Institute of Medical Research where the genotyping of the discovery sample was performed. We acknowledge Drs Neill R Graff-Radford, Dennis W Dickson and Ronald C Petersen for their key contribution in collecting the Mayo replication sample. One of the replication data sets was funded by the Alzheimer′s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904; RC2 AG036535). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Amorfix Life Sciences; AstraZeneca; Bayer HealthCare; BioClinica; Biogen Idec; Bristol-Myers Squibb Company; Eisai; Elan Pharmaceuticals; Eli Lilly and Company; F Hoffmann-La Roche and its affiliated company Genentech; GE Healthcare; Innogenetics, N.V.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace; Merck & Co.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer; Servier; Synarc; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer′s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129, K01 AG030514 and the Dana Foundation. Some sample collection was supported by the NIA National Cell Repository for Alzheimer's Disease (NCRAD; U24 AG021886) and additional support for data analysis was provided by R01 AG19771 and P30 AG10133.. We thank the following people for their contributions to the ADNI genotyping project: Tatiana Foroud, Kelley Faber, Li Shen, Steven G Potkin, Matthew J Huentelman, David W Craig, Sungeun Kim, Kwangsik Nho and Bryan M DeChairo.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Supplementary Information accompanies the paper on the Translational Psychiatry website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Kamboh, M., Demirci, F., Wang, X. et al. Genome-wide association study of Alzheimer's disease. Transl Psychiatry 2, e117 (2012). https://doi.org/10.1038/tp.2012.45
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2012.45
Keywords
This article is cited by
-
SSBlazer: a genome-wide nucleotide-resolution model for predicting single-strand break sites
Genome Biology (2024)
-
Systematic review: fluid biomarkers and machine learning methods to improve the diagnosis from mild cognitive impairment to Alzheimer’s disease
Alzheimer's Research & Therapy (2023)
-
Alzheimer risk gene product Pyk2 suppresses tau phosphorylation and phenotypic effects of tauopathy
Molecular Neurodegeneration (2022)
-
Evaluation of mild cognitive impairment genetic susceptibility risks in a Chinese population
BMC Psychiatry (2022)
-
Genomics and Functional Genomics of Alzheimer's Disease
Neurotherapeutics (2022)