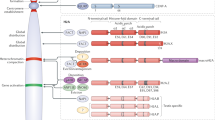
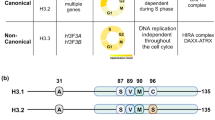
Key Points
-
Histone variants introduce small sequence variations into the eukaryotic epigenome to affect numerous aspects of chromatin structural organization, nucleosomal dynamics and transcription.
-
Histone variants have crucial roles during mammalian germ cell development and differentiation.
-
The exchange of histone variants is required to form normal patterns of zygotic and embryonic development.
-
Alterations in histone variant expression and/or activity have been directly linked to various forms of human disease, including cancer.
Abstract
Despite a conserved role for histones as general DNA packaging agents, it is now clear that another key function of these proteins is to confer variations in chromatin structure to ensure dynamic patterns of transcriptional regulation in eukaryotes. The incorporation of histone variants is particularly important to this process. Recent knockdown and knockout studies in various cellular systems, as well as direct mutational evidence from human cancers, now suggest a crucial role for histone variant regulation in processes as diverse as differentiation and proliferation, meiosis and nuclear reprogramming. In this Review, we provide an overview of histone variants in the context of their unique functions during mammalian germ cell and embryonic development, and examine the consequences of aberrant histone variant regulation in human disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Skene, P. J. & Henikoff, S. Histone variants in pluripotency and disease. Development 140, 2513–2524 (2013).
Chen, P., Zhao, J. & Li, G. Histone variants in development and diseases. J. Genet. Genom. 40, 355–365 (2013).
Filipescu, D., Szenker, E. & Almouzni, G. Developmental roles of histone H3 variants and their chaperones. Trends Genet. 29, 630–640 (2013).
Talbert, P. B. & Henikoff, S. Histone variants — ancient wrap artists of the epigenome. Nature Rev. Mol. Cell Biol. 11, 264–275 (2010).
Elsasser, S. J. et al. DAXX envelops a histone H3.3–H4 dimer for H3.3-specific recognition. Nature 491, 560–565 (2012).
Ranjan, A. et al. Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell 154, 1232–1245 (2013).
Yen, K., Vinayachandran, V. & Pugh, B. F. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell 154, 1246–1256 (2013).
Banaszynski, L. A., Allis, C. D. & Lewis, P. W. Histone variants in metazoan development. Dev. Cell 19, 662–674 (2010).
Drabent, B., Bode, C., Bramlage, B. & Doenecke, D. Expression of the mouse testicular histone gene H1t during spermatogenesis. Histochem. Cell Biol. 106, 247–251 (1996).
Martianov, I. et al. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc. Natl Acad. Sci. USA 102, 2808–2813 (2005).
Ishibashi, T. et al. H2A.Bbd: an X-chromosome-encoded histone involved in mammalian spermiogenesis. Nucleic Acids Res. 38, 1780–1789 (2010).
Govin, J. et al. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J. Cell Biol. 176, 283–294 (2007).
Zalensky, A. O. et al. Human testis/sperm-specific histone H2B (hTSH2B): molecular cloning and characterization. J. Biol. Chem. 277, 43474–43480 (2002).
Witt, O., Albig, W. & Doenecke, D. Testis-specific expression of a novel human H3 histone gene. Exp. Cell Res. 229, 301–306 (1996).
Schenk, R., Jenke, A., Zilbauer, M., Wirth, S. & Postberg, J. H3.5 is a novel hominid-specific histone H3 variant that is specifically expressed in the seminiferous tubules of human testes. Chromosoma 120, 275–285 (2011).
Bao, Y. et al. Nucleosomes containing the histone variant H2A.Bbd organize only 118 base pairs of DNA. EMBO J. 23, 3314–3324 (2004).
Syed, S. H. et al. The incorporation of the novel histone variant H2AL2 confers unusual structural and functional properties of the nucleosome. Nucleic Acids Res. 37, 4684–4695 (2009).
Soboleva, T. A. et al. A unique H2A histone variant occupies the transcriptional start site of active genes. Nature Struct. Mol. Biol. 19, 25–30 (2012).
Li, A. et al. Characterization of nucleosomes consisting of the human testis/sperm-specific histone H2B variant (hTSH2B). Biochemistry 44, 2529–2535 (2005).
Tachiwana, H. et al. Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T. Proc. Natl Acad. Sci. USA 107, 10454–10459 (2010).
Montellier, E. et al. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev. 27, 1680–1692 (2013). This study is the first to show that TSH2B exchange is required for protamine replacement during spermatogenesis.
Tolstorukov, M. Y. et al. Histone variant H2A.Bbd is associated with active transcription and mRNA processing in human cells. Mol. Cell 47, 596–607 (2012). This study illustrated that H2A.Bbd is enriched throughout active regions of the genome and forms a complex with both elongating RNA polymerase II and spliceosomal elements when stably expressed in HeLa cells.
Ioudinkova, E. S. et al. Distinct distribution of ectopically expressed histone variants H2A.Bbd and macroH2A in open and closed chromatin domains. PLoS ONE 7, e47157 (2012).
Hoyer-Fender, S., Costanzi, C. & Pehrson, J. R. Histone macroH2A1.2 is concentrated in the XY-body by the early pachytene stage of spermatogenesis. Exp. Cell Res. 258, 254–260 (2000).
Mahadevaiah, S. K. et al. Recombinational DNA double-strand breaks in mice precede synapsis. Nature Genet. 27, 271–276 (2001).
van der Heijden, G. W. et al. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nature Genet. 39, 251–258 (2007).
Fernandez-Capetillo, O. et al. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev. Cell 4, 497–508 (2003).
Greaves, I. K., Rangasamy, D., Devoy, M., Marshall Graves, J. A. & Tremethick, D. J. The X and Y chromosomes assemble into H2A.Z-containing facultative heterochromatin following meiosis. Mol. Cell. Biol. 26, 5394–5405 (2006).
Hammoud, S. S. et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460, 473–478 (2009). This paper is the first to use genome-wide sequencing to identify targets of histone retention in human sperm, with a specific emphasis on histone modifications.
Erkek, S. et al. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nature Struct. Mol. Biol. 20, 868–875 (2013).
Brykczynska, U. et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nature Struct. Mol. Biol. 17, 679–687 (2010).
Hammoud, S. S. et al. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum. Reprod. 26, 2558–2569 (2011).
Tanaka, M., Hennebold, J. D., Macfarlane, J. & Adashi, E. Y. A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development 128, 655–664 (2001).
Torres-Padilla, M. E., Bannister, A. J., Hurd, P. J., Kouzarides, T. & Zernicka-Goetz, M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int. J. Dev. Biol. 50, 455–461 (2006).
Akiyama, T., Suzuki, O., Matsuda, J. & Aoki, F. Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos. PLoS Genet. 7, e1002279 (2011).
Chang, C. C. et al. A maternal store of macroH2A is removed from pronuclei prior to onset of somatic macroH2A expression in preimplantation embryos. Dev. Biol. 278, 367–380 (2005).
Santenard, A. et al. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nature Cell Biol. 12, 853–862 (2010).
Orsi, G. A. et al. Drosophila Yemanuclein and HIRA cooperate for de novo assembly of H3.3-containing nucleosomes in the male pronucleus. PLoS Genet. 9, e1003285 (2013).
Loppin, B. et al. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437, 1386–1390 (2005). This work is the first to show that HIRA specifically incorporates H3.3 into paternal zygotic chromatin prior to the first round of DNA replication.
Costanzi, C., Stein, P., Worrad, D. M., Schultz, R. M. & Pehrson, J. R. Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development 127, 2283–2289 (2000).
Tang, M. C., Jacobs, S. A., Wong, L. H. & Mann, J. R. Conditional allelic replacement applied to genes encoding the histone variant H3.3 in the mouse. Genesis 51, 142–146 (2013).
Couldrey, C., Carlton, M. B., Nolan, P. M., Colledge, W. H. & Evans, M. J. A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Hum. Mol. Genet. 8, 2489–2495 (1999).
Bush, K. M. et al. Endogenous mammalian histone H3.3 exhibits chromatin-related functions during development. Epigenetics Chromatin 6, 7 (2013). This paper provides the first evidence that complete knockout of one gene copy encoding H3.3 ( H3f3b ) results in severe developmental deficits in mice.
Lin, C. J., Conti, M. & Ramalho-Santos, M. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development 140, 3624–3634 (2013).
Banaszynski, L. A. et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155, 107–120 (2013).
Hodl, M. & Basler, K. Transcription in the absence of histone H3.3. Curr. Biol. 19, 1221–1226 (2009). This study reveals that canonical H3.2 can fully compensate for H3.3 in D. melanogaster . These findings need to be verified in a mammalian model system.
Sakai, A., Schwartz, B. E., Goldstein, S. & Ahmad, K. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr. Biol. 19, 1816–1820 (2009).
Hodl, M. & Basler, K. Transcription in the absence of histone H3.2 and H3K4 methylation. Curr. Biol. 22, 2253–2257 (2012).
Eirin-Lopez, J. M., Gonzalez-Romero, R., Dryhurst, D., Ishibashi, T. & Ausio, J. The evolutionary differentiation of two histone H2A.Z variants in chordates (H2A.Z-1 and H2A.Z-2) is mediated by a stepwise mutation process that affects three amino acid residues. BMC Evol. Biol. 9, 31 (2009).
Faast, R. et al. Histone variant H2A.Z is required for early mammalian development. Curr. Biol. 11, 1183–1187 (2001).
Hu, G. et al. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 12, 180–192 (2013).
Creyghton, M. P. et al. H2AZ is enriched at Polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135, 649–661 (2008).
Wiedemann, S. M. et al. Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J. Cell Biol. 190, 777–791 (2010).
Chen, P. et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev. 27, 2109–2124 (2013).
Jin, C. et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nature Genet. 41, 941–945 (2009).
Nekrasov, M. et al. Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nature Struct. Mol. Biol. 19, 1076–1083 (2012). This paper shows that unstable nucleosomes located at the TSS are heterotypic with respect to H2A (that is, H2A.Z–H2A), which increases instability to an already unstable environment that is occupied by two variants (for example, H2A.Z–H3.3).
Costanzi, C. & Pehrson, J. R. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393, 599–601 (1998).
Buschbeck, M. et al. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nature Struct. Mol. Biol. 16, 1074–1079 (2009).
Gamble, M. J., Frizzell, K. M., Yang, C., Krishnakumar, R. & Kraus, W. L. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev. 24, 21–32 (2010).
Celeste, A. et al. Genomic instability in mice lacking histone H2AX. Science 296, 922–927 (2002).
Bassing, C. H. et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl Acad. Sci. USA 99, 8173–8178 (2002).
Celeste, A. et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114, 371–383 (2003).
Bassing, C. H. et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114, 359–370 (2003).
Izzo, A. et al. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep. 3, 2142–2154 (2013).
Lu, X. et al. Drosophila H1 regulates the genetic activity of heterochromatin by recruitment of Su(var)3-9. Science 340, 78–81 (2013).
Cao, K. et al. High-resolution mapping of H1 linker histone variants in embryonic stem cells. PLoS Genet. 9, e1003417 (2013).
Morin, R. D. et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood 122, 1256–1265 (2013).
Morin, R. D. et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011).
Okosun, J. et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nature Genet. 46, 176–181 (2014).
Pina, B. & Suau, P. Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev. Biol. 123, 51–58 (1987).
Toyama, B. H. et al. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 154, 971–982 (2013). This study uses metabolic pulse-chase labelling in rats to show that canonical H3 proteins (for example, H3.1) are highly stable in adult neural chromatin.
Hake, S. B. et al. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 281, 559–568 (2006).
Hake, S. B. & Allis, C. D. Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis”. Proc. Natl Acad. Sci. USA 103, 6428–6435 (2006).
Bernstein, E. & Hake, S. B. The nucleosome: a little variation goes a long way. Biochem. Cell Biol. 84, 505–517 (2006).
Michod, D. et al. Calcium-dependent dephosphorylation of the histone chaperone DAXX regulates H3.3 loading and transcription upon neuronal activation. Neuron 74, 122–135 (2012).
Najmabadi, H. et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478, 57–63 (2011).
Cox, S. G. et al. An essential role of variant histone H3.3 for ectomesenchyme potential of the cranial neural crest. PLoS Genet. 8, e1002938 (2012).
Bonisch, C. et al. H2A.Z.2.2 is an alternatively spliced histone H2A.Z variant that causes severe nucleosome destabilization. Nucleic Acids Res. 40, 5951–5964 (2012).
Santoro, S. W. & Dulac, C. The activity-dependent histone variant H2BE modulates the life span of olfactory neurons. eLife 1, e00070 (2012).
Vardabasso, C. et al. Histone variants: emerging players in cancer biology. Cell. Mol. Life Sci. 71, 379–404 (2013).
Graber, M. W., Schweinfest, C. W., Reed, C. E., Papas, T. S. & Baron, P. L. Isolation of differentially expressed genes in carcinoma of the esophagus. Ann. Surg. Oncol. 3, 192–197 (1996).
Schwartzentruber, J. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012). This work is the first to identify driver mutations that are specific to the histone variant H3.3 in human cancer.
Wu, G. et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nature Genet. 44, 251–253 (2012).
Khuong-Quang, D. A. et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 124, 439–447 (2012).
Lewis, P. W. et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013).
Chan, K. M. et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 27, 985–990 (2013). References 85 and 86 show the dominant negative effects of histone H3 mutations on the activity of PRC2 methyltransferase activity in paediatric gliomagenesis.
Goldberg, A. D. et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 (2010). This work provides the first genome-wide profiles of a histone variant (H3.3) in a mammalian system.
Elsaesser, S. J., Goldberg, A. D. & Allis, C. D. New functions for an old variant: no substitute for histone H3.3. Curr. Opin. Genet. Dev. 20, 110–117 (2010).
Kang, B. et al. Phosphorylation of H4 Ser47 promotes HIRA-mediated nucleosome assembly. Genes Dev. 25, 1359–1364 (2011).
Drane, P., Ouararhni, K., Depaux, A., Shuaib, M. & Hamiche, A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24, 1253–1265 (2010).
Lewis, P. W., Elsaesser, S. J., Noh, K. M., Stadler, S. C. & Allis, C. D. DAXX is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl Acad. Sci. USA 107, 14075–14080 (2010).
Jiao, Y. et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331, 1199–1203 (2011). This paper is the first to show that mutations in histone variant-associated chaperones are linked to human PanNETs.
Cheung, N. K. et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. J. Am. Med. Associ. 307, 1062–1071 (2012).
Ding, L. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012).
Garrick, D. et al. Loss of Atrx affects trophoblast development and the pattern of X-inactivation in extraembryonic tissues. PLoS Genet. 2, e58 (2006).
Michaelson, J. S., Bader, D., Kuo, F., Kozak, C. & Leder, P. Loss of Daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes Dev. 13, 1918–1923 (1999).
Baumann, C., Viveiros, M. M. & De La Fuente, R. Loss of maternal ATRX results in centromere instability and aneuploidy in the mammalian oocyte and pre-implantation embryo. PLoS Genet. 6, e1001137 (2010).
Wong, L. H. et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 20, 351–360 (2010).
Heaphy, C. M. et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science 333, 425 (2011).
Cao, Y. et al. Whole exome sequencing of insulinoma reveals recurrent T372R mutations in YY1. Nature Commun. 4, 2810 (2013).
Behjati, S. et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nature Genet. 45, 1479–1482 (2013). This recent paper identifies H3.3-specific driver mutations in chondroblastoma and giant cell tumours of bone, which indicates that H3.3 mutations probably result in a large variety of human cancers.
Broniscer, A. & Gajjar, A. Supratentorial high-grade astrocytoma and diffuse brainstem glioma: two challenges for the pediatric oncologist. Oncologist 9, 197–206 (2004).
Barrow, J. et al. Homozygous loss of ADAM3A revealed by genome-wide analysis of pediatric high-grade glioma and diffuse intrinsic pontine gliomas. Neuro. Oncol. 13, 212–222 (2011).
Bax, D. A. et al. A distinct spectrum of copy number aberrations in pediatric high-grade gliomas. Clin. Cancer Res. 16, 3368–3377 (2010).
Zarghooni, M. et al. Whole-genome profiling of pediatric diffuse intrinsic pontine gliomas highlights platelet-derived growth factor receptor α and poly (ADP-ribose) polymerase as potential therapeutic targets. J. Clin. Oncol. 28, 1337–1344 (2010).
Paugh, B. S. et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J. Clin. Oncol. 28, 3061–3068 (2010).
Margueron, R. & Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011).
Wagner, E. J. & Carpenter, P. B. Understanding the language of Lys36 methylation at histone H3. Nature Rev. Mol. Cell Biol. 13, 115–126 (2012).
Hock, H. A complex Polycomb issue: the two faces of EZH2 in cancer. Genes Dev. 26, 751–755 (2012).
Adam, S., Polo, S. E. & Almouzni, G. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 155, 94–106 (2013). This paper is the first to demonstrate an essential role for HIRA, which is an H3.3-specific chaperone, during periods of transcriptional recovery after DNA damage.
Black, B. E. & Cleveland, D. W. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell 144, 471–479 (2011).
Warburton, P. E. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 12, 617–626 (2004).
Howman, E. V. et al. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl Acad. Sci. USA 97, 1148–1153 (2000).
Mishra, P. K. et al. Misregulation of Scm3p/HJURP causes chromosome instability in Saccharomyces cerevisiae and human cells. PLoS Genet. 7, e1002303 (2011).
Tomonaga, T. et al. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 63, 3511–3516 (2003).
Biermann, K. et al. Gene expression profiling identifies new biological markers of neoplastic germ cells. Anticancer Res. 27, 3091–3100 (2007).
Wu, Q. et al. Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer 77, 407–414 (2012).
McGovern, S. L., Qi, Y., Pusztai, L., Symmans, W. F. & Buchholz, T. A. Centromere protein-A, an essential centromere protein, is a prognostic marker for relapse in estrogen receptor-positive breast cancer. Breast Cancer Res. 14, R72 (2012).
Li, Y. M., Liu, X. H., Cao, X. Z., Wang, L. & Zhu, M. H. [Expression of centromere protein A in hepatocellular carcinoma]. Zhonghua Bing Li Xue Za Zhi 36, 175–178 (in Chinese) (2007).
Li, Y. et al. shRNA-targeted centromere protein A inhibits hepatocellular carcinoma growth. PLoS ONE 6, e17794 (2011).
Hua, S. et al. Genomic analysis of estrogen cascade reveals histone variant H2A.Z associated with breast cancer progression. Mol. Systems Biol. 4, 188 (2008).
Dunican, D. S., McWilliam, P., Tighe, O., Parle-McDermott, A. & Croke, D. T. Gene expression differences between the microsatellite instability (MIN) and chromosomal instability (CIN) phenotypes in colorectal cancer revealed by high-density cDNA array hybridization. Oncogene 21, 3253–3257 (2002).
Rhodes, D. R. et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc. Natl Acad. Sci. USA 101, 9309–9314 (2004).
Zucchi, I. et al. Gene expression profiles of epithelial cells microscopically isolated from a breast-invasive ductal carcinoma and a nodal metastasis. Proc. Natl Acad. Sci. USA 101, 18147–18152 (2004).
Xu, Y. et al. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell 48, 723–733 (2012).
Greaves, I. K., Rangasamy, D., Ridgway, P. & Tremethick, D. J. H2A.Z contributes to the unique 3D structure of the centromere. Proc. Natl Acad. Sci. USA 104, 525–530 (2007).
Rangasamy, D., Berven, L., Ridgway, P. & Tremethick, D. J. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 22, 1599–1607 (2003).
Shia, W. J., Li, B. & Workman, J. L. SAS-mediated acetylation of histone H4 Lys16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev. 20, 2507–2512 (2006).
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 (1998).
Fernandez-Capetillo, O., Lee, A., Nussenzweig, M. & Nussenzweig, A. H2AX: the histone guardian of the genome. DNA Repair 3, 959–967 (2004).
Li, A. et al. Phosphorylation of histone H2A.X by DNA-dependent protein kinase is not affected by core histone acetylation, but it alters nucleosome stability and histone H1 binding. J. Biol. Chem. 285, 17778–17788 (2010).
Xiao, A. et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature 457, 57–62 (2009).
Zhu, F. et al. Phosphorylation of H2AX at Ser139 and a new phosphorylation site Ser16 by RSK2 decreases H2AX ubiquitination and inhibits cell transformation. Cancer Res. 71, 393–403 (2011).
Monni, O. & Knuutila, S. 11q deletions in hematological malignancies. Leuk. Lymphoma 40, 259–266 (2001).
Thirman, M. J. et al. Rearrangement of the MLL gene in acute lymphoblastic and acute myeloid leukemias with 11q23 chromosomal translocations. New Engl. J. Med. 329, 909–914 (1993).
Stankovic, T. et al. ATM mutations in sporadic lymphoid tumours. Leuk. Lymphoma 43, 1563–1571 (2002).
Parikh, R. A. et al. Loss of distal 11q is associated with DNA repair deficiency and reduced sensitivity to ionizing radiation in head and neck squamous cell carcinoma. Genes Chromosomes Cancer 46, 761–775 (2007).
Srivastava, N., Gochhait, S., Gupta, P. & Bamezai, R. N. Copy number alterations of the H2AFX gene in sporadic breast cancer patients. Cancer Genet. Cytogenet. 180, 121–128 (2008).
Sporn, J. C. & Jung, B. Differential regulation and predictive potential of macroH2A1 isoforms in colon cancer. Am. J. Pathol. 180, 2516–2526 (2012).
Sporn, J. C. et al. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene 28, 3423–3428 (2009).
Kapoor, A. et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature 468, 1105–1109 (2010). This study shows that mH2A is a critical chromatin factor that is associated with suppression of malignant melanoma development.
Novikov, L. et al. QKI-mediated alternative splicing of the histone variant macroH2A1 regulates cancer cell proliferation. Mol. Cell. Biol. 31, 4244–4255 (2011).
Li, X. et al. The atypical histone macroH2A1.2 interacts with HER-2 protein in cancer cells. J. Biol. Chem. 287, 23171–23183 (2012).
Zhang, R. et al. Formation of macroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8, 19–30 (2005).
Ratnakumar, K. et al. ATRX-mediated chromatin association of histone variant macroH2A1 regulates α-globin expression. Genes Dev. 26, 433–438 (2012).
Jullien, J., Astrand, C., Halley-Stott, R. P., Garrett, N. & Gurdon, J. B. Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation. Proc. Natl Acad. Sci. USA 107, 5483–5488 (2010).
Nashun, B., Akiyama, T., Suzuki, M. G. & Aoki, F. Dramatic replacement of histone variants during genome remodeling in nuclear-transferred embryos. Epigenet. 6, 1489–1497 (2011).
Chang, C. C. et al. Rapid elimination of the histone variant MacroH2A from somatic cell heterochromatin after nuclear transfer. Cell. Reprogramm. 12, 43–53 (2010).
Perez-Montero, S., Carbonell, A., Moran, T., Vaquero, A. & Azorin, F. The embryonic linker histone H1 variant of Drosophila, dBigH1, regulates zygotic genome activation. Dev. Cell 26, 578–590 (2013).
Barrero, M. J. et al. Macrohistone variants preserve cell identity by preventing the gain of H3K4me2 during reprogramming to pluripotency. Cell Rep. 3, 1005–1011 (2013).
Pasque, V. et al. Histone variant macroH2A marks embryonic differentiation in vivo and acts as an epigenetic barrier to induced pluripotency. J. Cell Sci. 125, 6094–6104 (2012).
Gaspar-Maia, A. et al. MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nature Commun. 4, 1565 (2013).
Schones, D. E. et al. Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887–898 (2008).
Luger, K. et al. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997).
Acknowledgements
The authors thank members of C.D.A.'s laboratory for providing critiques of the manuscript. Given the broad scope of this Review, they apologize for any relevant citations that might have been omitted. I.M., K.-M.N. and C.D.A. are partly supported by grants from the US National Institute of Mental Health (RO1 MH094698-01, P50 MH096890-01) and the Leukemia & Lymphoma Society (LLS-SCOR 7132–08). A.A.S. is supported by a Howard Hughes Medical Institute Damon Runyon Cancer Research Foundation fellowship (DRG-2185-14).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Nucleosomal
-
Pertaining to the nucleosome, which is the basic unit of chromatin that contains ~147 bp of DNA wrapped around a histone octamer (which is composed of two copies each of histone H3, H4, H2A and H2B, or variants thereof).
- Canonical histones
-
Archetypical histones of a given family to which all other histone proteins of that same family are compared.
- Imprinted loci
-
Genes that are expressed from only one of the two parental copies, the choice of which is dependent on the sex of the parent from which the gene was derived.
- Bivalent promoters
-
Regions of chromatin that have co-occurrence of histone H3 trimethylated at lysine 27 (H3K27me3) and H3K4me2 or H3K4me3 during embryonic development.
- Totipotent
-
Pertaining to the capacity of an undifferentiated cell to develop into any type of cell.
- Pronucleus
-
The haploid nucleus from a gamete.
- Hypomorphic
-
Pertaining to a mutant allele that does not completely eliminate the wild-type function of a gene and that gives a less severe phenotype than a loss-of-function mutant.
- Polycomb repressive complex
-
(PRC). An epigenetic regulator of gene expression that silences target genes by establishing a repressive chromatin state. PRC2 trimethylates histone H3 at lysine 27. This repressive histone modification is recognized by PRC1, which has ubiquitylating activity. As PRCs can maintain states of gene expression, they have key roles in cell fate maintenance and transitions during development.
- Trophectoderm
-
The outer layer of the blastocyst-stage embryo that gives rise to the trophoblast after implantation and that will provide the bulk of the extra-embryonic lineages of the placenta.
- Blastocyst
-
A pre-implantation embryonic stage that is characterized by the first definitive lineages. It contains a fluid-filled cavity (that is, the blastocoel), a focal cluster of cells from which the embryo develops (that is, the inner cell mass) and peripheral trophoblast cells (which form the placenta).
- Inner cell mass
-
A cluster of undifferentiated cells in the blastocyst, which give rise to the entire fetus and to some of its extra-embryonic (that is, placental) tissues.
- Gastrulation
-
The process by which the three primitive germ layers are formed in the early embryo; it is one of the first major differentiation events in development.
- Core transcription factor
-
A transcription factor that controls the expression of key pluripotency-related genes. Pluripotent cells are highly responsive to levels of these transcriptional regulators.
- Aneuploidy
-
The presence of an abnormal number of chromosomes.
- Blebbing
-
Formation of protrusions from a membrane. Nuclear blebs are caused by localized separation of fibres from the lamin meshwork.
- Bridging
-
Formation of abnormal connections between two nuclei, in which the nuclear membrane extends across two poorly separated or non-separated chromatin masses. Nuclear bridges are associated with failed and regressed cytokinesis.
- Haploinsufficiency
-
A genetic condition in a diploid organism in which a single functional copy of a gene fails to generate sufficient gene product, leading to an abnormal or diseased state.
Rights and permissions
About this article
Cite this article
Maze, I., Noh, KM., Soshnev, A. et al. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat Rev Genet 15, 259–271 (2014). https://doi.org/10.1038/nrg3673
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3673
This article is cited by
-
A sex-specific genome-wide association study of depression phenotypes in UK Biobank
Molecular Psychiatry (2023)
-
Histone 3.3-related chromatinopathy: missense variants throughout H3-3A and H3-3B cause a range of functional consequences across species
Human Genetics (2023)
-
Extra-nuclear histones: origin, significance and perspectives
Molecular and Cellular Biochemistry (2022)
-
Simultaneous disruption of PRC2 and enhancer function underlies histone H3.3-K27M oncogenic activity in human hindbrain neural stem cells
Nature Genetics (2021)
-
A dual role for H2A.Z.1 in modulating the dynamics of RNA polymerase II initiation and elongation
Nature Structural & Molecular Biology (2021)