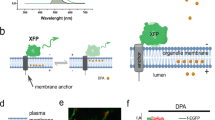
Abstract
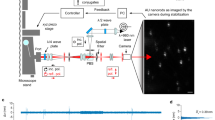
This protocol describes a sensitive approach to tracking the motion of membrane molecules such as lipids and proteins with molecular resolution in live cells. This technique makes use of fluorescent semiconductor nanocrystals, quantum dots (QDs), as a probe to detect membrane molecules of interest. The photostability and brightness of QDs allow them to be tracked at a single particle level for longer periods than previous fluorophores, such as fluorescent proteins and organic dyes. QDs are bound to the extracellular part of the object to be followed, and their movements can be recorded with a fluorescence microscope equipped with a spectral lamp and a sensitive cooled charge-coupled device camera. The experimental procedure described for neurons below takes about 45 min. This technique is applicable to various cultured cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Saxton, M.J. & Jacobson, K. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 26, 373–399 (1997).
Dahan, M. From analog to digital: exploring cell dynamics with single quantum dots. Histochem. Cell. Biol. 125, 451–456 (2006).
Geerts, H. et al. Nanovid tracking: a new automatic method for the study of mobility in living cells based on colloidal gold and video microscopy. Biophys. J. 52, 775–782 (1987).
Schnapp, B.J., Gelles, J. & Sheetz, M.P. Nanometer-scale measurements using video light microscopy. Cell. Motil. Cytoskeleton 10, 47–53 (1988).
Kusumi, A., Sako, Y. & Yamamoto, M. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys. J. 65, 2021–2040 (1993).
Meier, J., Vannier, C., Serge, A., Triller, A. & Choquet, D. Fast and reversible trapping of surface glycine receptors by gephyrin. Nat. Neurosci. 4, 253–260 (2001).
Borgdorff, A.J. & Choquet, D. Regulation of AMPA receptor lateral movements. Nature 417, 649–653 (2002).
Iino, R., Koyama, I. & Kusumi, A. Single molecule imaging of green fluorescent proteins in living cells: E-cadherin forms oligomers on the free cell surface. Biophys. J. 80, 2667–2677 (2001).
Schmidt, T., Schutz, G.J., Baumgartner, W., Gruber, H.J. & Schindler, H. Imaging of single molecule diffusion. Proc. Natl. Acad. Sci. USA 93, 2926–2929 (1996).
Tardin, C., Cognet, L., Bats, C., Lounis, B. & Choquet, D. Direct imaging of lateral movements of AMPA receptors inside synapses. EMBO J. 22, 4656–4665 (2003).
Schutz, G.J., Sonnleitner, M., Hinterdorfer, P. & Schindler, H. Single molecule microscopy of biomembranes (review). Mol. Membr. Biol. 17, 17–29 (2000).
Alivisatos, P. The use of nanocrystals in biological detection. Nat. Biotechnol. 22, 47–52 (2004).
Jaiswal, J.K. & Simon, S.M. Potentials and pitfalls of fluorescent quantum dots for biological imaging. Trends Cell Biol. 14, 497–504 (2004).
Michalet, X. et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science 307, 538–544 (2005).
Giepmans, B.N., Adams, S.R., Ellisman, M.H. & Tsien, R.Y. The fluorescent toolbox for assessing protein location and function. Science 312, 217–224 (2006).
Wu, X. et al. Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat. Biotechnol. 21, 41–46 (2003).
Pellegrino, T. et al. Quantum dot-based cell motility assay. Differentiation 71, 542–548 (2003).
Dubertret, B. et al. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science 298, 1759–1762 (2002).
Dahan, M. et al. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science 302, 442–445 (2003).
Groc, L. et al. Differential activity-dependent regulation of the lateral mobilities of AMPA and NMDA receptors. Nat. Neurosci. 7, 695–696 (2004).
Lidke, D.S. et al. Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nat. Biotechnol. 22, 198–203 (2004).
Pinaud, F., King, D., Moore, H.P. & Weiss, S. Bioactivation and cell targeting of semiconductor CdSe/ZnS nanocrystals with phytochelatin-related peptides. J. Am. Chem. Soc. 126, 6115–6123 (2004).
Charrier, C., Ehrensperger, M.V., Dahan, M., Levi, S. & Triller, A. Cytoskeleton regulation of glycine receptor number at synapses and diffusion in the plasma membrane. J. Neurosci. 26, 8502–8511 (2006).
Luccardini, C., Tribet, C., Vial, F., Marchi-Artzner, V. & Dahan, M. Size, charge, and interactions with giant lipid vesicles of quantum dots coated with an amphiphilic macromolecule. Langmuir 22, 2304–2310 (2006).
Giepmans, B.N., Deerinck, T.J., Smarr, B.L., Jones, Y.Z. & Ellisman, M.H. Correlated light and electron microscopic imaging of multiple endogenous proteins using Quantum dots. Nat. Methods 2, 743–749 (2005).
Sekine-Aizawa, Y. & Huganir, R.L. Imaging of receptor trafficking by using alpha-bungarotoxin-binding-site-tagged receptors. Proc. Natl. Acad. Sci. USA 101, 17114–17119 (2004).
Howarth, M., Takao, K., Hayashi, Y. & Ting, A.Y. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proc. Natl. Acad. Sci. USA 102, 7583–7588 (2005).
Fujiwara, T., Ritchie, K., Murakoshi, H., Jacobson, K. & Kusumi, A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 157, 1071–1081 (2002).
Bonneau, S., Cohen, L. & Dahan, M. A multiple target approach for single quantum dot tracking. Proc. IEEE Int. Symp. Biol. Imag. 664 (2004).
Bonneau, S., Dahan, M. & Cohen, L.D. Single quantum dot tracking based on perceptual grouping using minimal paths in a spatiotemporal volume. IEEE Trans. Image Process. 14, 1384–1395 (2005).
Courty, S., Luccardini, C., Bellaiche, Y., Cappello, G. & Dahan, M. Tracking individual kinesin motors in living cells using single quantum-dot imaging. Nano Lett. 6, 1491–1495 (2006).
Hanus, C., Ehrensperger, M.V. & Triller, A. Activity-dependent movements of postsynaptic scaffolds at inhibitory synapses. J. Neurosci. 26, 4586–4595 (2006).
Yao, J., Larson, D.R., Vishwasrao, H.D., Zipfel, W.R. & Webb, W.W. Blinking and nonradiant dark fraction of water-soluble quantum dots in aqueous solution. Proc. Natl. Acad. Sci. USA 102, 14284–14289 (2005).
Acknowledgements
We thank S. Bonneau and M.-V. Ehrensperger (Ecole Normale Supérieure, Paris, France) for writing the custom-made software for data analysis and T. Inoue and K. Mikoshiba (University of Tokyo, Japan) for the custom-made software TI Workbench and for valuable discussions on data analysis. We also thank Quantum Dot Corporation for providing the composition of the QD binding buffer. This work was supported by grants from INSERM, the CNRS, the Ministère de la Recherche and the FRM to S.L., M.D. and A.T. and Toyobo Biotechnology Foundation, Hayashi Memorial Foundation for Female Natural Scientists and the Japan Society for the Promotion of Science to H.B.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Video 1
Example of QD recording. (MOV 1574 kb)
Supplementary Video 2
Example of simultaneous QDs and calcium indicator imaging. (MOV 2325 kb)
Rights and permissions
About this article
Cite this article
Bannai, H., Lévi, S., Schweizer, C. et al. Imaging the lateral diffusion of membrane molecules with quantum dots. Nat Protoc 1, 2628–2634 (2006). https://doi.org/10.1038/nprot.2006.429
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.429
This article is cited by
-
Monitoring cell membrane recycling dynamics of proteins using whole-cell fluorescence recovery after photobleaching of pH-sensitive genetic tags
Nature Protocols (2022)
-
Effect of Ag2S-BSA nanoparticle size on 3T3 fibroblast cell line cytotoxicity
Journal of Nanoparticle Research (2020)
-
Limitations of Qdot labelling compared to directly-conjugated probes for single particle tracking of B cell receptor mobility
Scientific Reports (2017)
-
GABAA receptor dependent synaptic inhibition rapidly tunes KCC2 activity via the Cl−-sensitive WNK1 kinase
Nature Communications (2017)
-
Dynamic heterogeneity and non-Gaussian statistics for acetylcholine receptors on live cell membrane
Nature Communications (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.